[1]:
import os
os.environ[
"OMP_NUM_THREADS"
] = "64" # for jupyter.nersc.gov otherwise the notebook only uses 2 cores
[2]:
from pathlib import Path
import healpy as hp
import matplotlib.pyplot as plt
import numpy as np
import pymaster as nmt
from astropy.io import fits
%matplotlib inline
[3]:
plt.style.use("seaborn-talk")
[4]:
import pysm3 as pysm
import pysm3.units as u
[5]:
nside = 2048
lmax = 2048
[6]:
comp = "IQU"
[7]:
components = list(enumerate(comp))
components
[7]:
[(0, 'I'), (1, 'Q'), (2, 'U')]
[8]:
spectra_components = ["TT", "EE", "BB"]
change this to True if you want to run namaster on notebook
[9]:
namaster_on_nb = True
[10]:
datadir = Path("data/")
[11]:
proddatadir = Path("production-data") / "dust_gnilc" / "raw"
Setting the inputs¶
Dust maps¶
We use the 2015 GNILC intensity map from the 2nd planck release, as it encodes less contamination from CIB with 21.8’ resolution https://www.dropbox.com/s/hicocet83z31ob3/COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits?dl=0
for Q and U we adopt maps from the 3rd Planck release as they were optimized for polarization studies with 80’ reso.
[12]:
dust_varresI = datadir / "COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits"
[13]:
if not dust_varresI.exists():
!wget -O $dust_varresI https://portal.nersc.gov/project/cmb/pysm-data/dust_gnilc/inputs/COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits
Transform maps to double precision for computations
[14]:
I_planck_varres, h = hp.read_map(dust_varresI, dtype=np.float64, h=True)
Maps from the two releases are in different units MJy/sr the former, and K_CMB the latter, we therefore need to perform some conversion to uK_RJ.
[15]:
I_planck_varres <<= u.MJy / u.sr
I_planck_varres = I_planck_varres.to(
"uK_RJ", equivalencies=u.cmb_equivalencies(353 * u.GHz)
)
[16]:
output_nside = 8192
output_lmax = 2 * output_nside
Amplitude modulation¶
[17]:
modulate_amp = hp.alm2map(
hp.read_alm(
proddatadir / "gnilc_dust_temperature_modulation_alms_lmax768.fits.gz",
).astype(np.complex128),
nside=output_nside,
)
Injecting small scales to Spectral parameters¶
[18]:
tdfilename = "COM_CompMap_Dust-GNILC-Model-Temperature_2048_R2.01.fits"
tdfile = datadir / tdfilename
if not tdfile.exists():
!wget -O $tdfile https://irsa.ipac.caltech.edu/data/Planck/release_2/all-sky-maps/maps/component-maps/foregrounds/$tdfilename
bdfilename = "COM_CompMap_Dust-GNILC-Model-Spectral-Index_2048_R2.01.fits"
bdfile = datadir / bdfilename
if not bdfile.exists():
!wget -O $bdfile http://pla.esac.esa.int/pla/aio/product-action?MAP.MAP_ID=$bdfilename
[19]:
td = hp.read_map(tdfile, dtype=np.float64)
bd = hp.read_map(bdfile, dtype=np.float64)
[20]:
cltd = hp.anafast(td, use_pixel_weights=True, lmax=lmax)
clbd = hp.anafast(bd, use_pixel_weights=True, lmax=lmax)
cl = {"bd": clbd, "td": cltd}
dust_params = list(cl.keys())
ell = np.arange(lmax + 1)
[21]:
output_ell = np.arange(output_lmax + 1)
[22]:
from scipy.optimize import curve_fit
[22]:
def model(ell, A, gamma):
out = A * ell**gamma
out[:2] = 0
return out
[23]:
ell_fit_low = {"bd": 200, "td": 100}
ell_fit_high = {"bd": 400, "td": 400}
A_fit, gamma_fit, A_fit_std, gamma_fit_std = {}, {}, {}, {}
plt.figure(figsize=(25, 5))
for ii, pol in enumerate(dust_params):
plt.subplot(131 + ii)
xdata = np.arange(ell_fit_low[pol], ell_fit_high[pol])
ydata = cl[pol][xdata]
(A_fit[pol], gamma_fit[pol]), cov = curve_fit(model, xdata, ydata)
A_fit_std[pol], gamma_fit_std[pol] = np.sqrt(np.diag(cov))
plt.loglog(ell, cl[pol], label=" Anafast $C_\ell$")
plt.plot(
ell[ell_fit_low[pol] // 2 : ell_fit_high[pol] * 2],
model(
ell[ell_fit_low[pol] // 2 : ell_fit_high[pol] * 2],
A_fit[pol],
gamma_fit[pol],
),
label="model fit",
)
plt.axvline(
ell_fit_low[pol],
linestyle="--",
color="black",
label="$ \ell={} $".format(ell_fit_low[pol]),
)
plt.axvline(
ell_fit_high[pol],
linestyle="--",
color="gray",
label="$ \ell={} $".format(ell_fit_high[pol]),
)
plt.legend()
plt.grid()
plt.title(
f"{pol} spectrum for dust " + r"$\gamma_{fit}=$" + f"{gamma_fit[pol]:.2f}"
)
plt.ylabel("$ C_\ell $")
plt.xlabel(("$\ell$"))
plt.xlim(2, lmax)
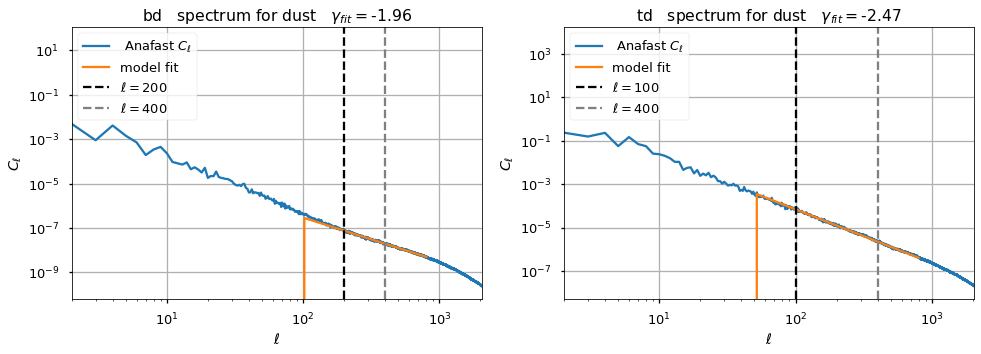
We inject smaller angular scales to the maps by extrapolating the power law fitted from the GNILC spectral parameter maps
Smaller angular scales are modulated similarly as the intensity map in pol tens formalism.
the multipoles where the fit is performed are different given the observed spectra . In any case we don’t fit beyond \(\ell=400\), which is consistent with the TT analysis above
given the fact that we inject smaller angular scales with a steeper spectral index than TT
\[\gamma_{\beta} = -1.96, \gamma_{Td} = -2.47, \gamma_{TT}= -1.29\]
we don’t expect to injecti small scale noise when rescaling at frequencies orders of magnitude lower or larger than the reference one ( 353 GHz).
[24]:
def sigmoid(x, x0, width, power=4):
"""Sigmoid function given start point and width
Parameters
----------
x : array
input x axis
x0 : float
value of x where the sigmoid starts (not the center)
width : float
width of the transition region in unit of x
power : float
tweak the steepness of the curve
Returns
-------
sigmoid : array
sigmoid, same length of x"""
return 1.0 / (1 + np.exp(-power * (x - x0 - width / 2) / width))
Small scales¶
[25]:
# filter small scales
small_scales_input_cl = [
1
* model(output_ell, A_fit[pol], gamma_fit[pol])
* (sigmoid(output_ell, ell_fit_high[pol], ell_fit_high[pol] / 10))
for pol in dust_params
]
names = ["beta", "Td"]
for name, each in zip(names, small_scales_input_cl):
hp.write_cl(
proddatadir / f"gnilc_dust_small_scales_{name}_cl_lmax{output_lmax}_2023.06.06.fits",
each,
dtype=np.complex128,
overwrite=True,
)
pysm.utils.add_metadata(
[proddatadir / f"gnilc_dust_small_scales_{name}_cl_lmax{output_lmax}_2023.06.06.fits"],
unit="K**2" if name == "Td" else "",
)
np.random.seed(777)
bd_ss_alm = hp.synalm(small_scales_input_cl[0], lmax=output_lmax)
np.random.seed(888)
td_ss_alm = hp.synalm(small_scales_input_cl[1], lmax=output_lmax)
bd_ss = hp.alm2map(bd_ss_alm, nside=output_nside)
td_ss = hp.alm2map(td_ss_alm, nside=output_nside)
bd_ss *= modulate_amp
td_ss *= modulate_amp
/tmp/ipykernel_265480/3589521984.py:2: RuntimeWarning: divide by zero encountered in power
out = A * ell ** gamma
[27]:
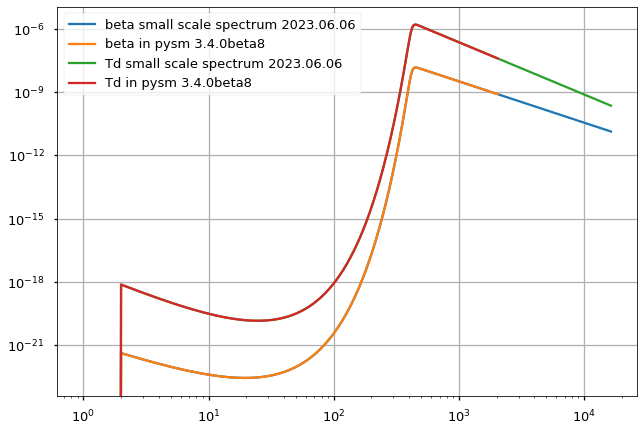
plt.loglog(small_scales_input_cl[0], label="beta small scale spectrum 2023.06.06")
plt.loglog(hp.read_cl("production-data/obsolete/dust_gnilc/pysm_3.4.0b8/gnilc_dust_small_scales_beta_cl_lmax16384.fits.gz"),
label="beta in pysm 3.4.0beta8")
plt.loglog(small_scales_input_cl[1], label="Td small scale spectrum 2023.06.06")
plt.loglog(hp.read_cl("production-data/obsolete/dust_gnilc/pysm_3.4.0b8/gnilc_dust_small_scales_Td_cl_lmax16384.fits.gz"),
label="Td in pysm 3.4.0beta8")
plt.legend()
plt.grid();
/global/homes/z/zonca/condanamaster2/lib/python3.8/site-packages/numpy/core/_asarray.py:102: ComplexWarning: Casting complex values to real discards the imaginary part
return array(a, dtype, copy=False, order=order)
/global/homes/z/zonca/condanamaster2/lib/python3.8/site-packages/numpy/core/_asarray.py:102: ComplexWarning: Casting complex values to real discards the imaginary part
return array(a, dtype, copy=False, order=order)
[ ]:
Large scales¶
[26]:
largescale_lmax = 1024
[27]:
alm_bd = hp.map2alm(bd, lmax=largescale_lmax, use_pixel_weights=True)
alm_td = hp.map2alm(td, lmax=largescale_lmax, use_pixel_weights=True)
sig_func = sigmoid(ell, x0=ell_fit_high["bd"], width=ell_fit_high["bd"] / 10)
bd_LS_alm = hp.almxfl(alm_bd, np.sqrt(1.0 - sig_func))
td_LS_alm = hp.almxfl(alm_td, np.sqrt(1.0 - sig_func))
if False: # do not generate large scales again
for name, alm in zip(names, [bd_LS_alm, td_LS_alm]):
hp.write_alm(
proddatadir
/ f"gnilc_dust_largescale_template_{name}_alm_nside{nside}_lmax{largescale_lmax:d}.fits",
alm,
lmax=largescale_lmax,
out_dtype=np.complex64,
overwrite=True,
)
pysm.utils.add_metadata(
[
proddatadir
/ f"gnilc_dust_largescale_template_beta_alm_nside{nside}_lmax{largescale_lmax:d}.fits"
],
coord="G",
unit="",
)
pysm.utils.add_metadata(
[
proddatadir
/ f"gnilc_dust_largescale_template_Td_alm_nside{nside}_lmax{largescale_lmax:d}.fits"
],
coord="G",
unit="K",
)
bd_ls = hp.alm2map(bd_LS_alm, nside=output_nside)
td_ls = hp.alm2map(td_LS_alm, nside=output_nside)
Final map¶
[28]:
bd_out = bd_ss + bd_ls
td_out = td_ss + td_ls
[29]:
# hp.write_map(
# datadir / f"gnilc_dust_beta_map_nside{output_nside}.fits",
# bd_out,
# dtype=np.float32,
# coord="G",
# column_units="",
# overwrite=True,
# )
[30]:
# hp.write_map(
# datadir / f"gnilc_dust_Td_map_nside{output_nside}.fits",
# td_out,
# dtype=np.float32,
# coord="G",
# column_units="K",
# overwrite=True,
# )
[31]:
cl_out = {
"bd": hp.anafast(bd_out, use_pixel_weights=True, lmax=lmax),
"td": hp.anafast(td_out, use_pixel_weights=True, lmax=lmax),
}
[32]:
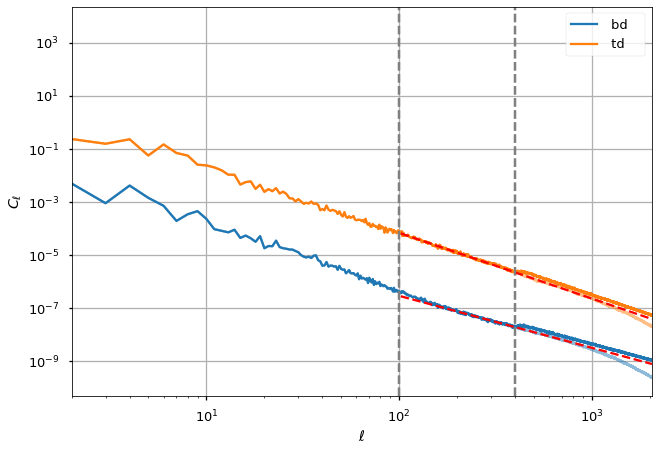
for ii, pol in enumerate(dust_params):
plt.loglog(ell, cl[pol], alpha=0.5, color="C%d" % ii)
plt.loglog(ell, cl_out[pol], label=f" {pol} ", color="C%d" % ii)
plt.legend()
plt.grid(True)
plt.plot(
ell[100:][2:],
model(ell[100:], A_fit[pol], gamma_fit[pol])[2:],
"--",
color="red",
)
plt.axvline(ell_fit_high[pol], linestyle="--", color="gray")
plt.axvline(100, linestyle="--", color="gray")
plt.ylabel("$ C_\ell $")
plt.xlabel(("$\ell$"))
# plt.xlim(350, 500 )
plt.xlim(2, lmax)
[33]:
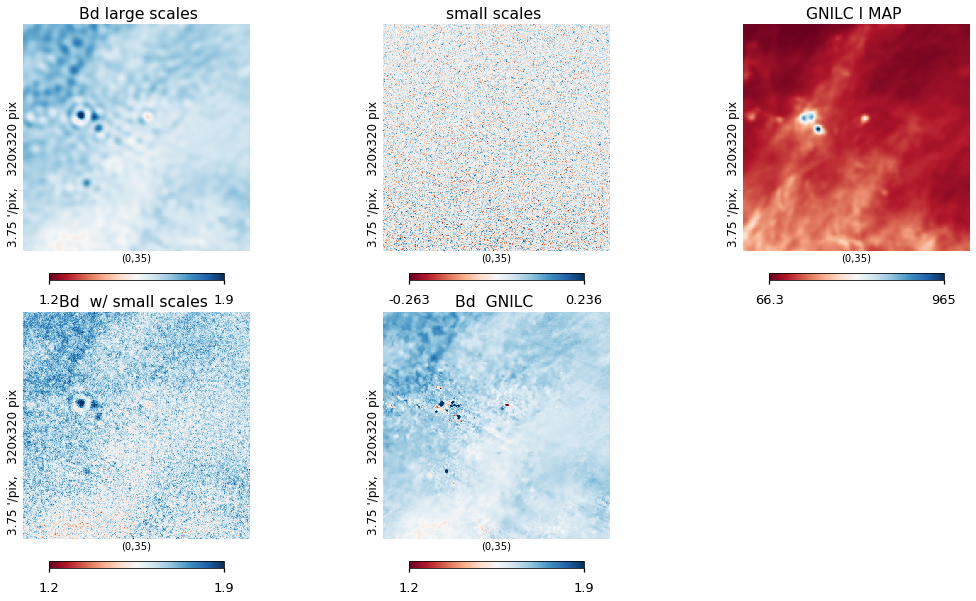
lat = 35
cm = plt.cm.RdBu
plt.figure(figsize=(15, 8))
hp.gnomview(
bd_out,
title="Bd w/ small scales ",
rot=[0, lat],
reso=3.75,
cmap=cm,
xsize=320,
sub=234,
min=1.2,
max=1.9,
)
hp.gnomview(
bd_ss, title="small scales ", rot=[0, lat], reso=3.75, xsize=320, cmap=cm, sub=232
)
hp.gnomview(
I_planck_varres,
title=" GNILC I MAP ",
rot=[0, lat],
reso=3.75,
cmap=cm,
xsize=320,
sub=233,
)
hp.gnomview(
(bd),
title=" Bd GNILC ",
rot=[0, lat],
reso=3.75,
xsize=320,
cmap=cm,
sub=235,
min=1.2,
max=1.9,
)
hp.gnomview(
bd_ls,
title=" Bd large scales ",
rot=[0, lat],
reso=3.75,
cmap=cm,
xsize=320,
sub=231,
min=1.2,
max=1.9,
)
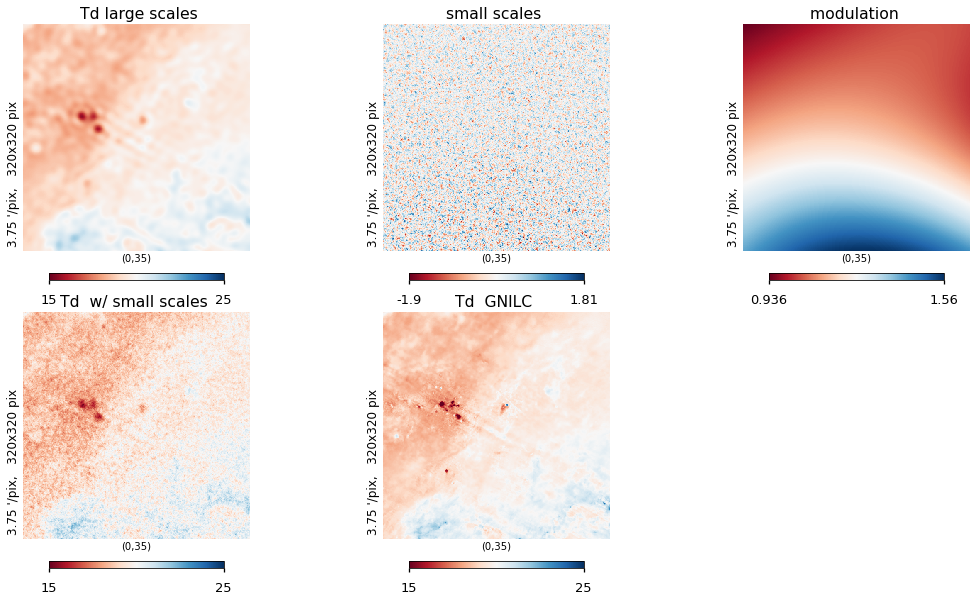
plt.figure(figsize=(15, 8))
hp.gnomview(
td_out,
title="Td w/ small scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
cmap=cm,
sub=234,
min=15,
max=25,
)
hp.gnomview(
td_ss, title="small scales ", rot=[0, lat], reso=3.75, xsize=320, sub=232, cmap=cm
)
hp.gnomview(
(modulate_amp),
title=" modulation ",
rot=[0, lat],
reso=3.75,
xsize=320,
cmap=cm,
sub=233,
)
hp.gnomview(
(td),
title=" Td GNILC ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=235,
cmap=cm,
min=15,
max=25,
)
hp.gnomview(
td_ls,
title=" Td large scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
cmap=cm,
sub=231,
min=15,
max=25,
)
Galactic plane fix¶
As in the templates, we replace the inner part of the galactic plane with the actual input data
[ ]:
templates_galplane_fix_mask = hp.read_map(
proddatadir / "gnilc_dust_galplane.fits.gz", 3
)
[ ]:
hp.mollview(templates_galplane_fix_mask)
[ ]:
for each in [td, bd]:
each[templates_galplane_fix_mask == 1] = 0
[39]:
if False: # do not generate galplane fix again
hp.write_map(
proddatadir / "gnilc_dust_beta_Td_galplane.fits.gz",
[bd, td],
fits_IDL=False,
overwrite=True,
coord="G",
column_names=["BETA", "TD"],
column_units=["", "K"],
dtype=np.float32,
)
[ ]: