GNILC Dust templates for PySM 3¶
Dust templates based on Planck GNILC maps in logarithmic polarization fraction formalism (logpoltens) with injection of simulated small scale fluctuations
This notebook implements and validates the model at Nside 2048, see the related notebooks:
Below a summary of the updates implemented on new dust models : - input templates from GNILC variable resolution and unires GNILC maps - two pivot scales ell1=110,ell2=2000 for small scale injection (see my previous posts ) - spectral indices for EE, BB coming from literature , Planck 2018 XI and for TT from Miville-Deschenes 2016 - we inject small scales with non-zero TE from
Planck 2018 XI - Inside the GAL097 mask (i.e. along the Gal. midplane) we don’t inject small scales, we simply keep the ones observed at high SNR by Planck. - Modulation of qu maps with a single map, p: i. to avoid modulation w/ negative values, ii. to preserve non-zero TE - we propose to modulate small scales as it has been done in pysm2(https://arxiv.org/pdf/1608.02841.pdf) , with a couple of differences: i. small scales are expected to be
injected with non-gaussian content (thanks to the logpoltens formalism); ii. split the sky with high reso pixels (nside=8) ; iii. use amplitude of E-mode spectra to derive the modulation template
[1]:
from pathlib import Path
import healpy as hp
import matplotlib.pyplot as plt
import numpy as np
import pymaster as nmt
from astropy.io import fits
%matplotlib inline
[2]:
import os
# for jupyter.nersc.gov otherwise the notebook only uses 2 cores
os.environ["OMP_NUM_THREADS"] = "64"
[3]:
hp.disable_warnings()
WARNING: AstropyDeprecationWarning: The disable_warnings function is deprecated and may be removed in a future version. [warnings]
[4]:
plt.style.use("seaborn-talk")
[5]:
import pysm3 as pysm
import pysm3.units as u
[6]:
nside = 2048
lmax = 2048
[7]:
comp = "IQU"
[8]:
components = list(enumerate(comp))
components
[8]:
[(0, 'I'), (1, 'Q'), (2, 'U')]
[9]:
spectra_components = ["TT", "EE", "BB", "TE"]
change this to True if you want to run namaster on notebook
[10]:
namaster_on_nb = True
[11]:
datadir = Path("data")
Setting the inputs¶
Dust maps¶
We use the 2015 GNILC intensity map from the 2nd planck release, as it encodes less contamination from CIB with 21.8’ resolution https://portal.nersc.gov/project/cmb/pysm-data/dust_gnilc/inputs/COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits
for Q and U we adopt maps from the 3rd Planck release as they were optimized for polarization studies with 80’ reso.
[12]:
gnilc_template = "varres"
dust_varresI = datadir / "COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits"
dust_varresP = (
datadir / f"COM_CompMap_IQU-thermaldust-gnilc-{gnilc_template}_2048_R3.00.fits"
)
[13]:
if not dust_varresI.exists():
!wget -O $dust_varresI https://portal.nersc.gov/project/cmb/pysm-data/dust_gnilc/inputs/COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits
--2023-02-10 08:12:26-- https://portal.nersc.gov/project/cmb/pysm-data/dust_gnilc/inputs/COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits
Resolving portal.nersc.gov (portal.nersc.gov)... 128.55.206.107, 128.55.206.110, 128.55.206.109, ...
Connecting to portal.nersc.gov (portal.nersc.gov)|128.55.206.107|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 402664320 (384M)
Saving to: ‘data/COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits’
data/COM_CompMap_Du 100%[===================>] 384.01M 66.5MB/s in 6.1s
2023-02-10 08:12:32 (63.1 MB/s) - ‘data/COM_CompMap_Dust-GNILC-F353_2048_21p8acm.fits’ saved [402664320/402664320]
[14]:
if not dust_varresP.exists():
!wget -O $dust_varresP http://pla.esac.esa.int/pla/aio/product-action?MAP.MAP_ID=COM_CompMap_IQU-thermaldust-gnilc-varres_2048_R3.00.fits
--2023-02-10 08:12:32-- http://pla.esac.esa.int/pla/aio/product-action?MAP.MAP_ID=COM_CompMap_IQU-thermaldust-gnilc-varres_2048_R3.00.fits
Resolving pla.esac.esa.int (pla.esac.esa.int)... 193.147.153.153
Connecting to pla.esac.esa.int (pla.esac.esa.int)|193.147.153.153|:80... connected.
HTTP request sent, awaiting response... 200
Length: unspecified [image/fits]
Saving to: ‘data/COM_CompMap_IQU-thermaldust-gnilc-varres_2048_R3.00.fits’
data/COM_CompMap_IQ [ <=> ] 1.88G 17.3MB/s in 1m 55s
2023-02-10 08:14:28 (16.6 MB/s) - ‘data/COM_CompMap_IQU-thermaldust-gnilc-varres_2048_R3.00.fits’ saved [2013275520]
Transform maps to double precision for computations
[15]:
m_planck_varres, h = hp.read_map(
dust_varresP, [c + "_STOKES" for c in comp], dtype=np.float64, h=True
)
I_planck_varres, h = hp.read_map(dust_varresI, dtype=np.float64, h=True)
Maps from the two releases are in different units MJy/sr the former, and K_CMB the latter, we therefore need to perform some conversion to uK_RJ.
[16]:
m_planck_varres <<= u.K_CMB
I_planck_varres <<= u.MJy / u.sr
m_planck_varres = m_planck_varres.to(
"uK_RJ", equivalencies=u.cmb_equivalencies(353 * u.GHz)
)
I_planck_varres = I_planck_varres.to(
"uK_RJ", equivalencies=u.cmb_equivalencies(353 * u.GHz)
)
then we are ready to combine both maps into one single TQU map.
[17]:
m_planck_varres[0] = I_planck_varres
del I_planck_varres
GAL080 Planck mask¶
we perform the monopole removal in a region outside the Galactic plane.
[18]:
planck_mask_filename = datadir / "HFI_Mask_GalPlane-apo2_2048_R2.00.fits"
if not planck_mask_filename.exists():
!wget -O $planck_mask_filename "http://pla.esac.esa.int/pla/aio/product-action?MAP.MAP_ID=HFI_Mask_GalPlane-apo2_2048_R2.00.fits"
--2023-02-10 08:14:56-- http://pla.esac.esa.int/pla/aio/product-action?MAP.MAP_ID=HFI_Mask_GalPlane-apo2_2048_R2.00.fits
Resolving pla.esac.esa.int (pla.esac.esa.int)... 193.147.153.153
Connecting to pla.esac.esa.int (pla.esac.esa.int)|193.147.153.153|:80... connected.
HTTP request sent, awaiting response... 200
Length: unspecified [image/fits]
Saving to: ‘data/HFI_Mask_GalPlane-apo2_2048_R2.00.fits’
data/HFI_Mask_GalPl [ <=> ] 1.50G 16.7MB/s in 96s
2023-02-10 08:16:33 (15.9 MB/s) - ‘data/HFI_Mask_GalPlane-apo2_2048_R2.00.fits’ saved [1610622720]
[19]:
planck_mask = hp.read_map(planck_mask_filename, ["GAL080"])
planck_mask = np.int_(np.ma.masked_not_equal(planck_mask, 0.0).mask)
fsky = planck_mask.sum() / planck_mask.size
print(f"masking {fsky} of the sky")
hp.mollview(planck_mask, title=f"Planck common galactic mask, {comp}")
masking 0.7912631829579672 of the sky

Monopole subtraction¶
Section 2.2 of Planck 2018 XII value reported: 0.13 MJy/sr
we subtract this term only to the I map for the pixels outside the Galactic plane mask.
[20]:
planck2018_monopole = (0.13 * u.MJy / u.sr).to(
u.uK_RJ, equivalencies=u.cmb_equivalencies(353 * u.GHz)
)
m_planck_varres[0][planck_mask] -= planck2018_monopole
We estimate how many pixels have I< P after we subtract the monopole
[21]:
maskmono = m_planck_varres[0] ** 2 < m_planck_varres[1] ** 2 + m_planck_varres[2] ** 2
print(
f"{maskmono.sum() } pixels out of { maskmono.size} expected to be NaNs in Log Pol Tens maps "
)
5 pixels out of 50331648 expected to be NaNs in Log Pol Tens maps
[22]:
plt.figure(figsize=(20, 5))
for i_pol, pol in components:
hp.mollview(
m_planck_varres[i_pol],
title="Planck-GNILC 2058/2018 dust " + pol,
sub=131 + i_pol,
unit=m_planck_varres.unit,
min=-300,
max=300,
)
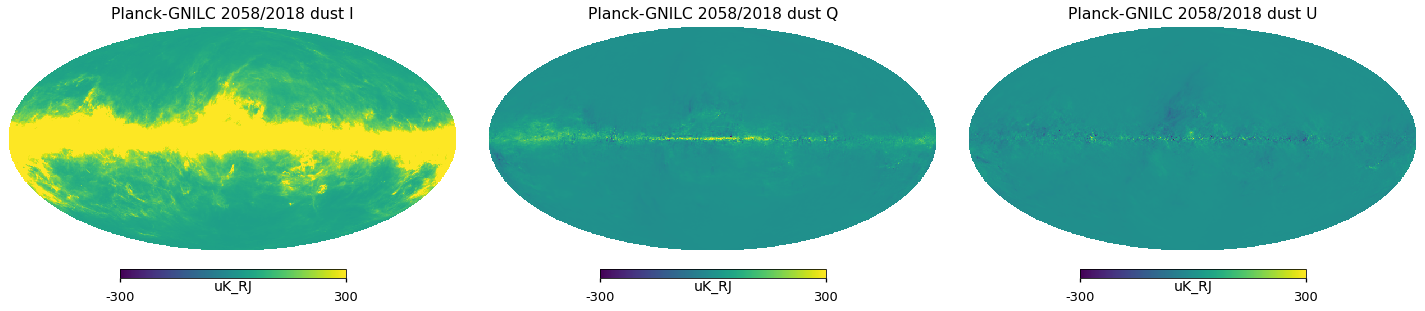
Transform maps to Poltens quantities¶
[23]:
def map_to_log_pol_tens(m):
P = np.sqrt(m[1] ** 2 + m[2] ** 2)
log_pol_tens = np.empty_like(m)
log_pol_tens[0] = np.log(m[0] ** 2 - P ** 2) / 2.0
log_pol_tens[1:] = m[1:] / P * np.log((m[0] + P) / (m[0] - P)) / 2.0
return log_pol_tens
def log_pol_tens_to_map(log_pol_tens):
P = np.sqrt(log_pol_tens[1] ** 2 + log_pol_tens[2] ** 2)
m = np.empty_like(log_pol_tens)
exp_i = np.exp(log_pol_tens[0])
m[0] = exp_i * np.cosh(P)
m[1:] = log_pol_tens[1:] / P * exp_i * np.sinh(P)
return m
def sigmoid(x, x0, width, power=4):
"""Sigmoid function given start point and width
Parameters
----------
x : array
input x axis
x0 : float
value of x where the sigmoid starts (not the center)
width : float
width of the transition region in unit of x
power : float
tweak the steepness of the curve
Returns
-------
sigmoid : array
sigmoid, same length of x"""
return 1.0 / (1 + np.exp(-power * (x - x0 - width / 2) / width))
[24]:
log_pol_tens_varres = map_to_log_pol_tens(m_planck_varres.value)
/tmp/ipykernel_7767/1566809701.py:4: RuntimeWarning: invalid value encountered in log
log_pol_tens[0] = np.log(m[0] ** 2 - P ** 2) / 2.0
/tmp/ipykernel_7767/1566809701.py:5: RuntimeWarning: invalid value encountered in log
log_pol_tens[1:] = m[1:] / P * np.log((m[0] + P) / (m[0] - P)) / 2.0
Checking NaNs on the Poltens map
[25]:
print(
f"{np.isnan(log_pol_tens_varres[0]).sum() } pixels out of { maskmono.size} are NaNs in Log Pol Tens maps "
)
5 pixels out of 50331648 are NaNs in Log Pol Tens maps
[26]:
for i in range(3):
log_pol_tens_varres[i, np.isnan(log_pol_tens_varres[i])] = np.nanmedian(
log_pol_tens_varres[i]
)
Set all the NaNs to the map median value
[27]:
assert np.isnan(log_pol_tens_varres).sum() == 0
[28]:
for i_pol, pol in components:
hp.mollview(
log_pol_tens_varres[i_pol],
title="Log Pol tensor " + pol,
sub=131 + i_pol,
unit=m_planck_varres.unit,
)
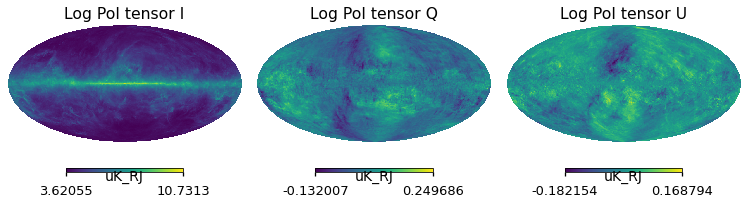
[29]:
from scipy.optimize import curve_fit
[30]:
def model(ell, A, gamma):
out = A * ell ** gamma
return out
[31]:
def run_anafast(m, lmax):
clanaf = hp.anafast(m, lmax=lmax)
cl = {}
cl["TT"] = clanaf[0]
cl["EE"] = clanaf[1]
cl["BB"] = clanaf[2]
cl["TE"] = clanaf[3]
ell = np.arange(lmax + 1)
cl_norm = ell * (ell + 1) / np.pi / 2
cl_norm[0] = 1
return ell, cl_norm, cl
def run_namaster(m, mask, lmax, nlbins):
"""Compute C_ell with NaMaster
Parameters
----------
m : numpy array
T only or TQU HEALPix map
mask : numpy array
mask, 1D, 0 for masked pixels,
needs to have same Nside of the input map
lmax : int
maximum ell of the spherical harmonics transform
Returns
-------
ell : numpy array
array of ell from 0 to lmax (length lmax+1)
cl_norm : numpy array
ell (ell+1)/2pi factor to turn C_ell into D_ell
first element is set to 1
cl : dict of numpy arrays
dictionary of numpy arrays with all components
of the spectra, for now only II, EE, BB, no
cross-spectra
"""
nside = hp.npix2nside(len(mask))
# b = nmt.NmtBin.from_nside_linear(nside, 16)
# leff = b.get_effective_ells()
binning = nmt.NmtBin(nside=nside, nlb=nlbins, lmax=lmax, is_Dell=False)
cl = {}
if len(m) == 3:
f_0 = nmt.NmtField(mask, [m[0]])
f_2 = nmt.NmtField(
mask, m[1:].copy(), purify_b=True
) # NaMaster masks the map in-place
cl_namaster = nmt.compute_full_master(f_2, f_2, binning)
cl["EE"] = np.concatenate([[0, 0], cl_namaster[0]])
cl["BB"] = np.concatenate([[0, 0], cl_namaster[3]])
cl_namaster = nmt.compute_full_master(f_0, f_2, binning)
cl["TE"] = np.concatenate([[0, 0], cl_namaster[0]])
elif m.ndim == 1:
m = m.reshape((1, -1))
f_0 = nmt.NmtField(mask, [m[0]])
cl_namaster_I = nmt.compute_full_master(f_0, f_0, binning)
cl["TT"] = np.concatenate([[0, 0], cl_namaster_I[0]])
ell = np.concatenate([[0, 1], binning.get_effective_ells()])
cl_norm = ell * (ell + 1) / np.pi / 2
cl_norm[0] = 1
return ell, cl_norm, cl
[32]:
print("run anafast on full sky ")
ell, cl_norm, cl = run_anafast(log_pol_tens_varres, lmax)
run anafast on full sky
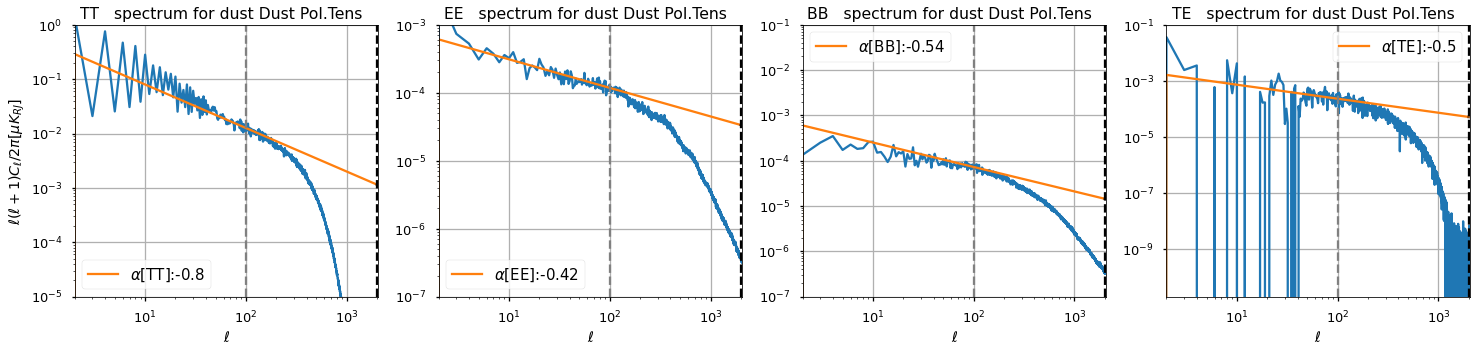
(New) employing spectral indices from literature¶
https://arxiv.org/pdf/1801.04945.pdf and https://www.aanda.org/articles/aa/pdf/2016/09/aa28503-16.pdf
2 pivotal scales
ell1=110andell2=800non zero TE spectrum
[33]:
ell_fit_low = {"TT": 50, "EE": 50, "BB": 50, "TE": 50}
ell_fit_high = {"TT": 100, "EE": 100, "BB": 100, "TE": 100}
gamma_fit2 = {"TT": -0.8, "EE": -0.42, "BB": -0.54, "TE": -0.50}
A_fit, gamma_fit, A_fit_std, gamma_fit_std = {}, {}, {}, {}
plt.figure(figsize=(25, 5))
A_fit2 = {}
smallscales = []
ell_pivot = 2000
for ii, pol in enumerate(spectra_components):
plt.subplot(141 + ii)
xdata = np.arange(ell_fit_low[pol], ell_fit_high[pol])
ydata = xdata * (xdata + 1) / np.pi / 2 * cl[pol][xdata]
(A_fit[pol], gamma_fit[pol]), cov = curve_fit(model, xdata, ydata)
A_fit2[pol] = np.fabs(A_fit[pol]) * ell_fit_high[pol] ** (
gamma_fit[pol] - gamma_fit2[pol]
)
plt.loglog(ell, ell * (ell + 1) / np.pi / 2 * cl[pol])
scaling = model(ell[:ell_pivot], A_fit2[pol], gamma_fit2[pol])
scaling[:2] = 0
plt.plot(ell[:ell_pivot], scaling, label=r"$\alpha$" + f"[{pol}]:{gamma_fit2[pol]}")
smallscales.append(scaling)
plt.axvline(ell_fit_high[pol], linestyle="--", color="gray")
plt.axvline(ell_pivot, linestyle="--", color="k")
plt.grid()
plt.title(f"{pol} spectrum for dust Dust Pol.Tens ")
plt.xlabel(("$\ell$"))
plt.xlim(2, lmax)
plt.legend(fontsize=15)
for ii, pol in enumerate(spectra_components):
# we change the EE and BB power laws
xdata = np.arange(ell_fit_high[pol], ell.size)
ydata = xdata * (xdata + 1) / np.pi / 2 * cl[pol][xdata]
(A_fit[pol], gamma_fit[pol]), cov = curve_fit(model, xdata, ydata)
plt.subplot(141 + ii)
if pol == "TE":
A_fit2[pol] = A_fit2[pol] * ell_pivot ** (gamma_fit2[pol] - gamma_fit2["TE"])
scaling = model(ell[ell_pivot:], A_fit2[pol], gamma_fit2["TE"])
plt.plot(
ell[ell_pivot:],
scaling,
linewidth=3,
alpha=0.4,
color="k",
)
smallscales[ii] = np.concatenate([smallscales[ii], scaling])
else:
A_fit2[pol] = A_fit2[pol] * ell_pivot ** (gamma_fit2[pol] - gamma_fit2["TT"])
scaling = model(ell[ell_pivot:], A_fit2[pol], gamma_fit2["TT"])
plt.plot(
ell[ell_pivot:],
scaling,
linewidth=3,
alpha=0.4,
color="k",
)
smallscales[ii] = np.concatenate([smallscales[ii], scaling])
plt.subplot(141)
plt.ylabel("$\ell(\ell+1)C_\ell/2\pi [\mu K_{RJ}]$")
plt.ylim(1e-5, 1e0)
plt.subplot(142)
plt.ylim(1e-7, 1e-3)
plt.subplot(143)
plt.ylim(1e-7, 1e-3)
plt.subplot(143)
plt.ylim(1e-7, 1e-1)
/tmp/ipykernel_7767/3409733669.py:2: RuntimeWarning: divide by zero encountered in power
out = A * ell ** gamma
/tmp/ipykernel_7767/3409733669.py:2: RuntimeWarning: divide by zero encountered in power
out = A * ell ** gamma
/tmp/ipykernel_7767/3409733669.py:2: RuntimeWarning: divide by zero encountered in power
out = A * ell ** gamma
/tmp/ipykernel_7767/3409733669.py:2: RuntimeWarning: divide by zero encountered in power
out = A * ell ** gamma
[33]:
(1e-07, 0.1)
[34]:
cl_ss = [
smallscales[ii] * sigmoid(ell, ell_fit_high[pol], ell_fit_high[pol] / 10) / cl_norm
for ii, pol in enumerate(spectra_components)
]
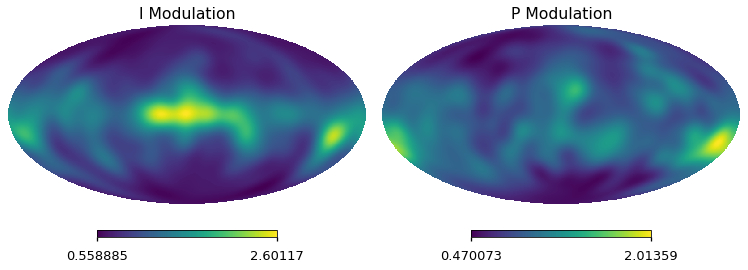
(NEW) Modulating intensity and polarization small scales¶
we will produce a modulation map at a very low reso (~ 11.3 degrees), to save memory we consider
nside=512we downgrade the
iqumap atnside=512for the same reason as abovewe considering the center of pixels at
nside=8we estimate the TT and EE power spectra in a circle of 11.3 deg apodized by 5 deg and centered in each
nside=8pixelthe modulation value in the patch \(p\) is given by :
\[A _p = \sqrt{\frac{C_{\ell=80, p}} {C_{ell=80,full}} }\]notice that in a given pixel we actually take an average value as the circles of neighbouring pixels overlap, for this reason we have to divide the modulation map by the map encoding the effective number of estimates
we apply a smoothing w/ a gaussian beam at 11.5 deg, to avoid sharp transitions .
[35]:
nsidepatches = 8
centers = np.vstack(
hp.pix2vec(ipix=np.arange(hp.nside2npix(nsidepatches)), nside=nsidepatches)
).T
mout = hp.ud_grade(log_pol_tens_varres, nside_out=512)
fit_model = lambda x, Ad, alpha: Ad * (x / 100) ** (alpha)
tmod = np.zeros(hp.nside2npix(hp.get_nside(mout)))
pmod = np.zeros(hp.nside2npix(hp.get_nside(mout)))
n_eff = np.zeros_like(mout[0])
def bin_cell(cell, dig):
cb = []
errb = []
for i in np.unique(dig):
msk = dig == i
cb.append(cell[msk].mean())
errb.append(cell[msk].std())
return np.array(cb), np.array(errb)
def bin_ell(ells, dig):
lb = []
dl = []
for i in np.unique(dig):
msk = dig == i
lb.append(ells[msk].mean())
dl.append((ells[msk].max() - ells[msk].min()) / 2)
return np.array(lb), np.array(dl)
for ipix, c in enumerate(centers):
patch = np.zeros_like(mout[0])
maskpixs = hp.query_disc(nside=hp.get_nside(mout), vec=c, radius=np.radians(11.3))
patch[maskpixs] = 1
apo_patch = nmt.mask_apodization(patch, 5, apotype="C2")
fsky = apo_patch.sum() / apo_patch.size
if ipix % 100 == 0:
print(ipix)
ellp, norm, clp = run_anafast(m=mout * apo_patch, lmax=512)
# bin the anafast spectra
digi = np.digitize(ellp, np.logspace(1, 2.5, 24))
dtt, errtt = bin_cell((clp["TT"]) / fsky, digi)
dee, erree = bin_cell((clp["EE"]) / fsky, digi)
lb, delta_l = bin_ell(ellp, digi)
# fit power law parameters for ell<100 for EE and 80<ell<200 for TT
lmask = (lb) < 100
lmaskt = np.logical_and((lb) < 200, lb > 80)
param_ee, _ = curve_fit(
fit_model, ydata=dee[lmask], xdata=lb[lmask], sigma=erree[lmask]
)
param_tt, _ = curve_fit(
fit_model, ydata=dtt[lmaskt], xdata=lb[lmaskt], sigma=errtt[lmaskt]
)
# assign modulation in the circle patch
l_ = 80
pmod += np.sqrt(fit_model(l_, *param_ee) / cl["EE"][l_]) * apo_patch
tmod += np.sqrt(fit_model(l_, *param_tt) / cl["TT"][l_]) * apo_patch
n_eff += apo_patch
0
100
200
300
400
500
600
700
[36]:
psm = hp.smoothing(pmod / n_eff, fwhm=np.radians(11.5))
tsm = hp.smoothing(tmod / n_eff, fwhm=np.radians(11.5))
hp.mollview(psm, sub=122, title="P Modulation")
hp.mollview(tsm, sub=121, title="I Modulation")
[37]:
hp.write_map(
datadir / "dust_gnilc_modulation_nside512.fits",
[tsm, psm],
column_units="",
coord="G",
column_names=["TEMPERATURE", "POLARISATION"],
dtype=np.float32,
overwrite=True,
)
[38]:
output_nside = 2048
output_lmax = output_nside
lmax = output_lmax
ell = np.arange(output_lmax + 1)
cl_norm = ell * (ell + 1) / np.pi / 2
cl_norm[:1] = 1
[39]:
np.random.seed(8192)
[40]:
log_ss = hp.synfast(
cl_ss,
lmax=lmax,
new=True,
nside=output_nside,
)
[41]:
polarization_mod = hp.ud_grade(psm, nside_out=output_nside)
temperature_mod = hp.ud_grade(tsm, nside_out=output_nside)
[42]:
log_ss_modulated = log_ss.copy()
log_ss_modulated[0] = log_ss[0] * temperature_mod
log_ss_modulated[1:] = log_ss[1:] * polarization_mod
[43]:
alm_log_pol_tens_fullsky = hp.map2alm(
log_pol_tens_varres, lmax=output_lmax, use_pixel_weights=True
)
ii_LS_alm = np.empty_like(alm_log_pol_tens_fullsky)
for ii, pol in enumerate(spectra_components[:-1]):
sig_func = sigmoid(ell, x0=ell_fit_high[pol], width=ell_fit_high[pol] / 10)
ii_LS_alm[ii] = hp.almxfl(alm_log_pol_tens_fullsky[ii], (1.0 - sig_func) ** 0.2)
log_ls = hp.alm2map(ii_LS_alm, nside=output_nside)
ii_outmap = log_ls + log_ss_modulated # * planck_mask + (1- planck_mask)* log_ss
small scales included only outside the GAL097 planck mask¶
Employ observed data inside the Gal. Plane
[44]:
galplane_mask = hp.read_map(planck_mask_filename, ["GAL097"])
[45]:
output_map = log_pol_tens_to_map(ii_outmap)
output_map = galplane_mask * output_map + (1 - galplane_mask) * m_planck_varres.value
[46]:
hp.write_map(
datadir / "dust_gnilc_template_nside2048.fits",
output_map,
column_units="uK_RJ",
coord="G",
dtype=np.float32,
overwrite=True,
)
[47]:
plt.figure(figsize=(15, 5))
hp.gnomview(
log_ss_modulated[0],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=152,
title="small-scales poltens i ",
)
hp.gnomview(
log_ls[0],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=153,
title="large-scales poltens i ",
)
hp.gnomview(
ii_outmap[0], reso=3.75, xsize=320, rot=[30, -58], sub=151, title="coadded i "
)
hp.gnomview(
log_pol_tens_varres[0],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=155,
title="input i ",
)
hp.gnomview(
temperature_mod, reso=3.75, xsize=320, rot=[30, -58], sub=154, title="modulation i "
)
plt.figure(figsize=(15, 5))
hp.gnomview(
log_ss_modulated[1],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=152,
title="small-scales poltens q",
)
hp.gnomview(
log_ls[1],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=153,
title="large-scales poltens q",
)
hp.gnomview(
ii_outmap[1], reso=3.75, xsize=320, rot=[30, -58], sub=151, title="coadded q "
)
hp.gnomview(
log_pol_tens_varres[1],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=155,
title="input q ",
)
hp.gnomview(
polarization_mod,
reso=3.75,
xsize=320,
rot=[30, -58],
sub=154,
title="modulation p ",
)
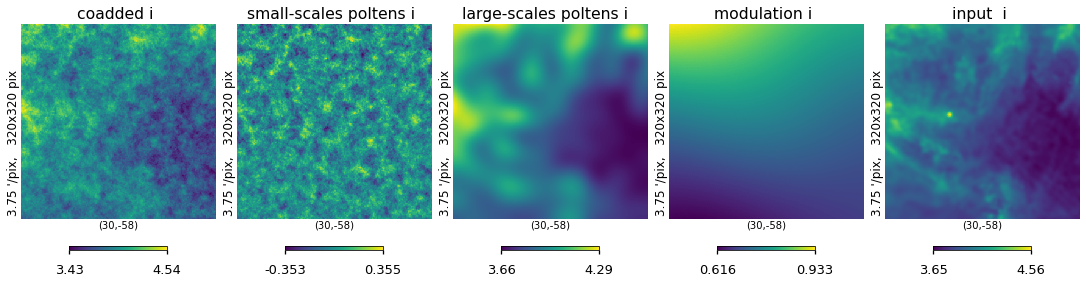
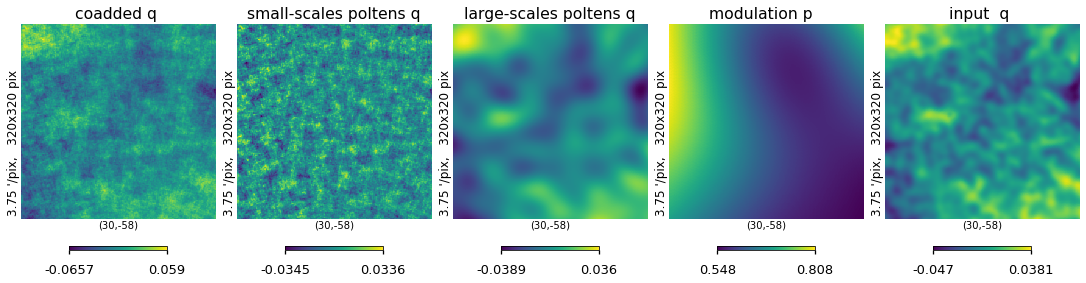
[48]:
del (
log_ls,
ii_LS_alm,
log_ss,
log_ss_modulated,
alm_log_pol_tens_fullsky,
)
del temperature_mod, polarization_mod
UDgrade to accelerate the Namaster computation¶
[49]:
output_map512 = hp.ud_grade(output_map, nside_out=512)
input_map512 = hp.ud_grade(m_planck_varres.value, nside_out=512)
[50]:
bkmaskfile = datadir / "bk14_mask_gal_n0512.fits"
if not bkmaskfile.exists():
!wget -O $bkmaskfile http://bicepkeck.org/BK14_datarelease/bk14_mask_gal_n0512.fits
maskbk = hp.read_map(bkmaskfile, verbose=False)
idx = np.where((maskbk < 0) | (~np.isfinite(maskbk)))
maskbk[idx] = 0
--2023-02-10 10:09:46-- http://bicepkeck.org/BK14_datarelease/bk14_mask_gal_n0512.fits
Resolving bicepkeck.org (bicepkeck.org)... 140.247.151.131
Connecting to bicepkeck.org (bicepkeck.org)|140.247.151.131|:80... connected.
HTTP request sent, awaiting response... 200 OK
Length: 12591360 (12M) [image/fits]
Saving to: ‘data/bk14_mask_gal_n0512.fits’
data/bk14_mask_gal_ 100%[===================>] 12.01M 3.26MB/s in 4.4s
2023-02-10 10:09:51 (2.72 MB/s) - ‘data/bk14_mask_gal_n0512.fits’ saved [12591360/12591360]
WARNING: AstropyDeprecationWarning: "verbose" was deprecated in version 1.15.0 and will be removed in a future version. [warnings]
[51]:
aoa = datadir / "weights_aoa_spsat_apo_gal_512.fits"
if not aoa.exists():
!wget -O $aoa "https://portal.nersc.gov/project/cmb/pysm-data/dust_gnilc/inputs/weights_aoa_spsat_apo_gal_512.fits"
--2023-02-10 10:09:52-- https://portal.nersc.gov/project/cmb/pysm-data/dust_gnilc/inputs/weights_aoa_spsat_apo_gal_512.fits
Resolving portal.nersc.gov (portal.nersc.gov)... 128.55.206.112, 128.55.206.110, 128.55.206.106, ...
Connecting to portal.nersc.gov (portal.nersc.gov)|128.55.206.112|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 12591360 (12M)
Saving to: ‘data/weights_aoa_spsat_apo_gal_512.fits’
data/weights_aoa_sp 100%[===================>] 12.01M --.-KB/s in 0.1s
2023-02-10 10:09:53 (83.6 MB/s) - ‘data/weights_aoa_spsat_apo_gal_512.fits’ saved [12591360/12591360]
[52]:
mask_aoa = hp.read_map(aoa, verbose=False)
WARNING: AstropyDeprecationWarning: "verbose" was deprecated in version 1.15.0 and will be removed in a future version. [warnings]
[53]:
planck_masks = {
"GAL099": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL099"]), nside_out=512),
"GAL097": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL097"]), nside_out=512),
"GAL090": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL090"]), nside_out=512),
"GAL080": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL080"]), nside_out=512),
"GAL070": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL070"]), nside_out=512),
"GAL060": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL060"]), nside_out=512),
"GAL040": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL040"]), nside_out=512),
"GAL020": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL020"]), nside_out=512),
}
[54]:
planck_masks_2048 = {
"GAL099": hp.read_map(planck_mask_filename, ["GAL099"]),
"GAL097": hp.read_map(planck_mask_filename, ["GAL097"]),
"GAL090": hp.read_map(planck_mask_filename, ["GAL090"]),
"GAL080": hp.read_map(planck_mask_filename, ["GAL080"]),
"GAL070": hp.read_map(planck_mask_filename, ["GAL070"]),
"GAL060": hp.read_map(planck_mask_filename, ["GAL060"]),
"GAL040": hp.read_map(planck_mask_filename, ["GAL040"]),
"GAL020": hp.read_map(planck_mask_filename, ["GAL020"]),
}
[55]:
ell, cl_norm, clo = run_anafast(output_map512, lmax=2048)
ell, cl_norm, cli = run_anafast(input_map512, lmax=2048)
[56]:
colors = plt.cm.cividis(np.linspace(0, 1, len(planck_masks.keys()) + 1))
[57]:
bins = {
"GAL099": 5,
"GAL097": 5,
"GAL090": 5,
"GAL080": 5,
"GAL070": 15,
"GAL060": 15,
"GAL040": 25,
"GAL020": 25,
}
plt.figure(figsize=(15, 10))
import os
# for jj, k in enumerate([ "GAL080","GAL070","GAL040","GAL020" ]) :
for jj, k in enumerate(planck_masks.keys()):
# fspectra = datadir / f"dust_gnilc_varres_no_monopole_gal{k[3:]}_spectra.npz"
fspectra = (
datadir / f"dust_gnilc_{gnilc_template}_no_monopole_gal{k[3:]}_spectra.npz"
)
print(k)
if os.path.exists(fspectra):
input_ell = np.load(fspectra)["ell"]
cl_in = {kk: np.load(fspectra)[kk] for kk in spectra_components + ["TE"]}
else:
input_ell, cl_norm, cl_in = run_namaster(
m=input_map512, mask=planck_masks[k], lmax=1300, nlbins=bins[k]
)
np.savez(
fspectra,
ell=input_ell,
TT=cl_in["TT"],
EE=cl_in["EE"],
BB=cl_in["BB"],
TE=cl_in["TE"],
)
fspectra = (
datadir
/ f"dust_gnilc_{gnilc_template}_pysm_circles_planck_gal{k[3:]}_spectra.npz"
)
if os.path.exists(fspectra):
output_ell = np.load(fspectra)["ell"]
cl_out = {kk: np.load(fspectra)[kk] for kk in spectra_components + ["TE"]}
else:
# output_ell, cl_norm, cl_out = run_namaster(m = output_map, mask =planck_masks[k] ,lmax=3000 , nlbins = bins[k] )
output_ell, cl_norm, cl_out = run_namaster(
m=output_map512, mask=planck_masks[k], lmax=1300, nlbins=bins[k]
)
np.savez(
fspectra,
ell=output_ell,
TT=cl_out["TT"],
EE=cl_out["EE"],
BB=cl_out["BB"],
TE=cl_out["TE"],
)
for ii, pol in enumerate(["TT", "EE", "BB", "TE"]):
plt.subplot(2, 2, ii + 1)
if jj == 0:
plt.title(pol)
plt.loglog(
ell,
ell ** 2 * (clo[pol]),
label="Small-Scales ",
color=colors[0],
)
plt.loglog(
ell,
ell ** 2 * (cli[pol]),
":",
label="GNILC map ",
color=colors[0],
alpha=0.5,
)
plt.loglog(output_ell, output_ell ** 2 * (cl_out[pol]), color=colors[jj])
# ,label =f"{k}")
plt.loglog(
input_ell,
input_ell ** 2 * (cl_in[pol]),
":",
color=colors[jj + 1],
alpha=0.5,
)
plt.subplot(221)
plt.legend()
plt.ylabel("$ D_\ell [\mu K_{RJ}]$")
plt.ylim(1e0, 2e6)
plt.xlim(2, 1e3)
plt.subplot(222)
plt.ylim(1e-1, 2e3)
plt.xlim(2, 1e3)
sm = plt.cm.ScalarMappable(
cmap=plt.cm.cividis_r, norm=plt.Normalize(vmin=20.0, vmax=100)
)
cb = plt.colorbar(sm)
cb.set_label(r" $f_{sky}$ [%]", rotation="90", fontsize=16)
plt.subplot(223)
plt.ylabel("$ D_\ell [\mu K_{RJ}]$")
plt.xlabel(("$\ell$"))
plt.ylim(1e-1, 2e3)
plt.xlim(2, 1e3)
plt.subplot(224)
plt.ylim(1e-2, 1e5)
plt.xlim(2, 1e3)
plt.xlabel(("$\ell$"))
sm = plt.cm.ScalarMappable(
cmap=plt.cm.cividis_r, norm=plt.Normalize(vmin=20.0, vmax=100)
)
cb = plt.colorbar(sm)
cb.set_label(r" $f_{sky}$ [%]", rotation="90", fontsize=16)
plt.tight_layout()
GAL099
GAL097
GAL090
GAL080
GAL070
GAL060
GAL040
GAL020
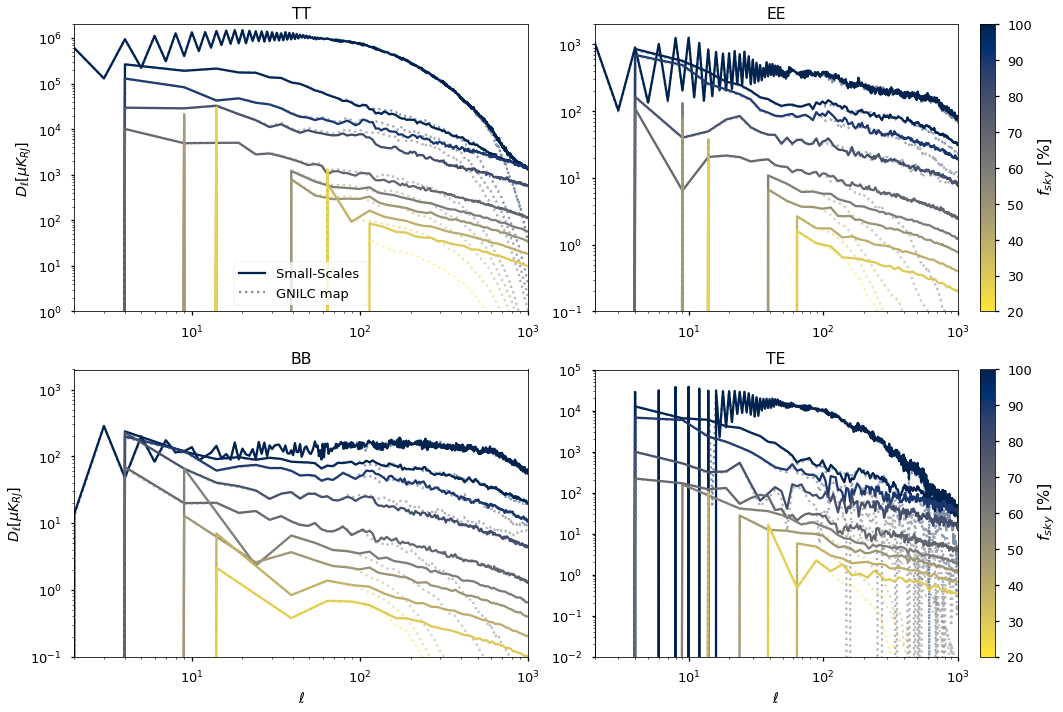
[58]:
# plt.figure(figsize=(15,10 ))
import os
for jj, k in enumerate(planck_masks.keys()):
fspectra = datadir / f"dust_gnilc_varres_pysm_circles_planck_gal{k[3:]}_spectra.npz"
if os.path.exists(fspectra):
# print("read Namaster spectra ")
output_ell = np.load(fspectra)["ell"]
cl_out = {kk: np.load(fspectra)[kk] for kk in spectra_components + ["TE"]}
else:
# print("computing spectra")
output_ell, cl_norm, cl_out = run_namaster(
m=output_map512, mask=planck_masks[k], lmax=1300, nlbins=bins[k]
)
np.savez(
fspectra,
ell=output_ell,
TT=cl_out["TT"],
EE=cl_out["EE"],
BB=cl_out["BB"],
TE=cl_out["TE"],
)
if jj == 0:
plt.plot(ell, (clo["EE"] / clo["BB"]), color=colors[0], alpha=1)
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
plt.xlim(200, 1e3)
plt.ylim(0, 3)
sm = plt.cm.ScalarMappable(
cmap=plt.cm.cividis_r, norm=plt.Normalize(vmin=20.0, vmax=100)
)
cb = plt.colorbar(sm)
cb.set_label(r" $f_{sky}$ [%]", rotation="90", fontsize=16)
plt.xlabel(("$\ell$"))
plt.ylabel(("E-to-B ratio"))
plt.title(gnilc_template)
plt.tight_layout()
/tmp/ipykernel_7767/3560805298.py:27: RuntimeWarning: invalid value encountered in true_divide
plt.plot(ell, (clo["EE"] / clo["BB"]), color=colors[0], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_7767/3560805298.py:28: RuntimeWarning: invalid value encountered in true_divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
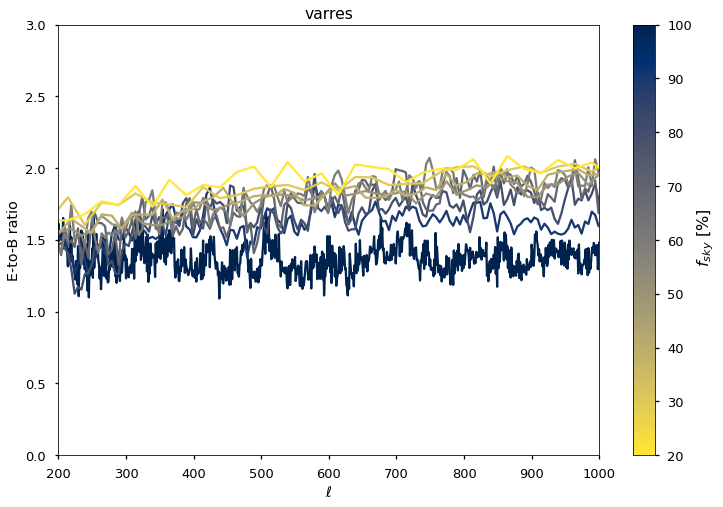
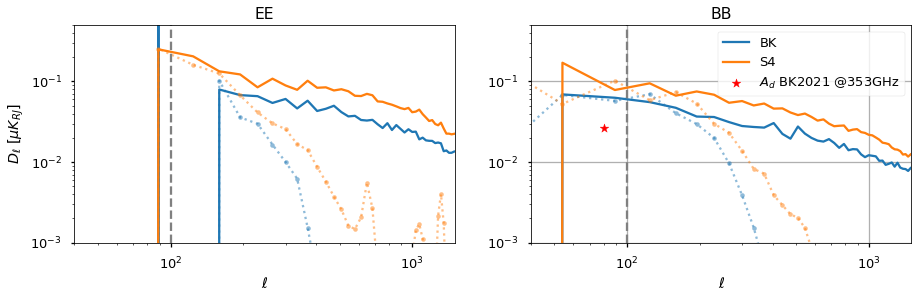
we further validate E and B spectra also within the BK and Southern Hole patches¶
[59]:
def ClBB(mask, mapp):
map1 = mapp.copy()
b = nmt.NmtBin.from_nside_linear(hp.get_nside(mask), 35)
f_2 = nmt.NmtField(mask, map1, purify_b=True)
cl_22 = nmt.compute_full_master(f_2, f_2, b)
ell_arr = b.get_effective_ells()
return (ell_arr, cl_22[3], cl_22[0])
[60]:
import matplotlib
import matplotlib.pyplot as plt
from matplotlib import rc
[61]:
i_dic = {}
(ells, clBB_i, clEE_i) = ClBB(mask_aoa, input_map512[1:, :])
i_dic["S4"] = [clBB_i, clEE_i]
(ells, clBB_i, clEE_i) = ClBB(maskbk, input_map512[1:, :])
i_dic["BK"] = [clBB_i, clEE_i]
[62]:
o_dic = {}
(oells, clBB_o, clEE_o) = ClBB(mask_aoa, output_map512[1:, :])
o_dic["S4"] = [clBB_o, clEE_o]
(oells, clBB_o, clEE_o) = ClBB(maskbk, output_map512[1:, :])
o_dic["BK"] = [clBB_o, clEE_o]
[63]:
bk = (np.sqrt(4.4) * u.uK_CMB).to(
"uK_RJ", equivalencies=u.cmb_equivalencies(353 * u.GHz)
)
[64]:
plt.figure(figsize=(15, 4))
plt.subplot(121)
plt.title("EE")
plt.ylabel("$ D_\ell\,\, [\mu K_{RJ}]$")
plt.xlabel(("$\ell$"))
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["BK"][1] / (2.0 * np.pi),
".",
alpha=0.5,
color="C0",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["BK"][1] / (2.0 * np.pi),
label="BK",
color="C0",
)
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["S4"][1] / (2.0 * np.pi),
".",
alpha=0.5,
color="C1",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["S4"][1] / (2.0 * np.pi),
label="Southern-Hole",
color="C1",
)
plt.loglog()
plt.ylim(1e-3, 5e-1)
plt.xlim(40, 1.5e3)
plt.axvline(ell_fit_high[pol], linestyle="--", color="gray")
plt.axvline(ell_pivot, linestyle="--", color="k")
plt.subplot(122)
plt.title("BB")
plt.xlabel(("$\ell$"))
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["BK"][0] / (2.0 * np.pi),
".",
alpha=0.5,
color="C0",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["BK"][0] / (2.0 * np.pi),
label="BK",
color="C0",
)
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["S4"][0] / (2.0 * np.pi),
".",
alpha=0.5,
color="C1",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["S4"][0] / (2.0 * np.pi),
label="S4",
color="C1",
)
plt.scatter(
[80.0], [(bk ** 2).value], color="r", marker="*", label=r"$A_d$ BK2021 @353GHz"
)
plt.loglog()
plt.ylim(1e-3, 5e-1)
plt.xlim(40, 1.5e3)
plt.axvline(ell_fit_high[pol], linestyle="--", color="gray")
plt.axvline(ell_pivot, linestyle="--", color="k")
plt.legend()
plt.grid()
Check polarization¶
[65]:
get_polfrac = lambda x: np.sqrt(x[1] ** 2 + x[2] ** 2) / x[0]
[66]:
Pout = get_polfrac(output_map)
Pin = get_polfrac(m_planck_varres.value)
logpin = np.log10(Pin)
logpout = np.log10(Pout)
[67]:
planck_masks_bool = {
k: np.ma.masked_greater(m, 0).mask for k, m in planck_masks_2048.items()
}
[68]:
mask_aoa2048 = hp.ud_grade(mask_aoa, nside_out=2048)
mask_bk2048 = hp.ud_grade(maskbk, nside_out=2048)
[69]:
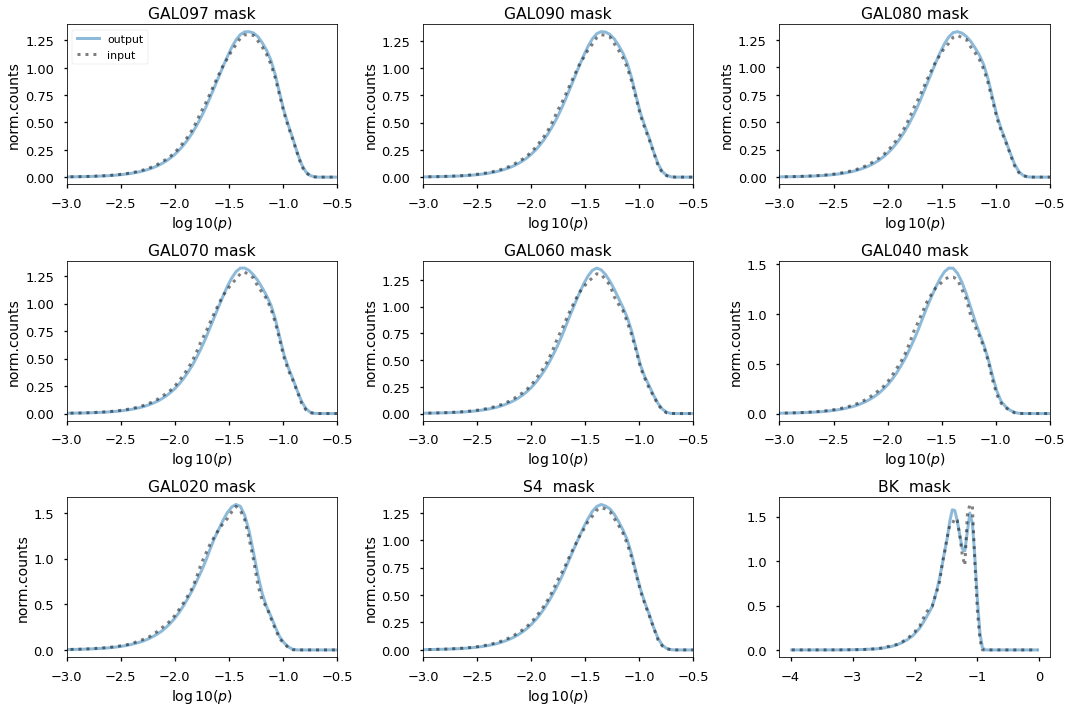
plt.figure(figsize=(15, 10))
for jj, pm in enumerate(planck_masks_bool.items()):
k = pm[0]
msk = pm[1]
print(k)
if k == "GAL099":
continue
plt.subplot(3, 3, jj)
h, edg = np.histogram(logpout[msk], bins=np.linspace(-4, 0, 100), density=True)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, color="C0", alpha=0.5, label="output")
h, edg = np.histogram(logpin[msk], density=True, bins=np.linspace(-4, 0, 100))
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, alpha=0.5, color="k", linestyle=":", label="input")
plt.ylabel("norm.counts", fontsize=14)
plt.xlabel(r"$\log10( p )$", fontsize=14)
plt.title(k + " mask")
plt.xlim(-3, -0.5)
plt.tight_layout()
plt.subplot(331)
plt.legend(fontsize=11, loc="upper left")
plt.subplot(339)
h, edg = np.histogram(
logpout[np.ma.masked_greater(mask_bk2048, 0).mask],
bins=np.linspace(-4, 0, 100),
density=True,
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, color="C0", alpha=0.5, label="output")
h, edg = np.histogram(
logpin[np.ma.masked_greater(mask_bk2048, 0).mask],
density=True,
bins=np.linspace(-4, 0, 100),
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, alpha=0.5, color="k", linestyle=":", label="input")
plt.title("BK mask")
plt.subplot(338)
h, edg = np.histogram(
logpout[np.ma.masked_greater(mask_aoa2048, 0).mask],
bins=np.linspace(-4, 0, 100),
density=True,
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, color="C0", alpha=0.5, label="output")
h, edg = np.histogram(
logpin[np.ma.masked_greater(mask_aoa2048, 0).mask],
density=True,
bins=np.linspace(-4, 0, 100),
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, alpha=0.5, color="k", linestyle=":", label="input")
plt.ylabel("norm.counts", fontsize=14)
plt.xlabel(r"$\log10( p )$", fontsize=14)
plt.title("S4 mask")
plt.xlim(-3, -0.5)
GAL099
GAL097
GAL090
GAL080
GAL070
GAL060
GAL040
GAL020
[69]:
(-3.0, -0.5)
[ ]: