Haslam/WMAP Synchrotron templates for PySM 3¶
Synchrotron templates based on Haslam for Temperature and WMAP for polarization in logarithmic polarization fraction formalism (logpoltens) with injection of simulated small scale fluctuations
This new implementation of small scale injection will homogeneize the synchrotron models to the latest updates implemented on dust : - input templates the same as before: HASLAM (rescaled to 23GHz and WMAP K-band, smoothed at 2deg) - two pivot scales ell1=36,ell2=400 for small scale injection - spectral indices for EE, BB coming from literature , Planck 2018 : i. alpha_TT=alpha_TE= -1 ii. alpha_EE = -0.84 iii. alpha_BB= -0.78 Notice that
for TT and TE the value is justified by what actually see in the spectra of input maps, see the plot in cell [31]. - small scales have with non-zero TE - Modulation of qu maps with a single map, p: i. to avoid modulation w/ negative values, ii. to preserve non-zero TE - we propose to modulate polarization small scales as it has been done in pysm2(https://arxiv.org/pdf/1608.02841.pdf) , with a couple of differences: i. small scales are expected to be injected with non-gaussian content
(thanks to the logpoltens formalism); ii. split the sky with high reso pixels (nside=8) ; iii. use amplitude of E-mode spectra to derive the modulation template
Related notebooks¶
Simplified version of this notebook that generates small scales up to :math:ell_{max}=16384` <synchrotron_template_logpoltens_generate_artifacts.ipynb>`__
Notebook that generates small scales for the spectral index :math:beta` <synchrotron_beta.ipynb>`__
Notebook that generates the template for curvature for ``s7` <synchrotron_curvature.ipynb>`__
Notebooks that create maps at different resolutions for templates, for spectral index and for curvature
[1]:
import os
os.environ[
"OMP_NUM_THREADS"
] = "64" # for jupyter.nersc.gov otherwise the notebook only uses 2 cores
output_files = []
from datetime import date
today = date.today()
version = today.strftime("%Y.%m.%d")
version
[1]:
'2023.02.24'
[2]:
from pathlib import Path
import healpy as hp
import matplotlib.pyplot as plt
import numpy as np
# import pymaster as nmt
from astropy.io import fits
%matplotlib inline
plt.style.use("seaborn-talk")
/tmp/ipykernel_17579/3930590641.py:11: MatplotlibDeprecationWarning: The seaborn styles shipped by Matplotlib are deprecated since 3.6, as they no longer correspond to the styles shipped by seaborn. However, they will remain available as 'seaborn-v0_8-<style>'. Alternatively, directly use the seaborn API instead.
plt.style.use("seaborn-talk")
[3]:
hp.disable_warnings()
WARNING: AstropyDeprecationWarning: The disable_warnings function is deprecated and may be removed in a future version. [warnings]
[4]:
import pysm3 as pysm
import pysm3.units as u
from pysm3.utils import add_metadata
[5]:
nside = 512
lmax = 3 * nside
[6]:
comp = "IQU"
[7]:
components = list(enumerate(comp))
components
[7]:
[(0, 'I'), (1, 'Q'), (2, 'U')]
[8]:
spectra_components = ["TT", "EE", "BB", "TE"]
change this to True if you want to run namaster on notebook
[9]:
namaster_on_nb = True
[10]:
datadir = Path("synch_data")
[11]:
proddir = Path("production-data") / "synch" / "raw"
[12]:
imapfile = datadir / "haslam408_dsds_Remazeilles2014.fits"
qumapfile = datadir / "wmap_band_iqumap_r9_9yr_K_v5.fits"
[13]:
if not imapfile.exists():
!wget -O $imapfile https://lambda.gsfc.nasa.gov/data/foregrounds/haslam_2014/haslam408_dsds_Remazeilles2014.fits
[14]:
if not qumapfile.exists():
!wget -O $qumapfile https://lambda.gsfc.nasa.gov/data/map/dr5/skymaps/9yr/raw/wmap_band_iqumap_r9_9yr_K_v5.fits
[15]:
imap = hp.read_map(imapfile)
qumap = hp.read_map(qumapfile, field=[1, 2])
Setting the inputs¶
Synchrotron maps¶
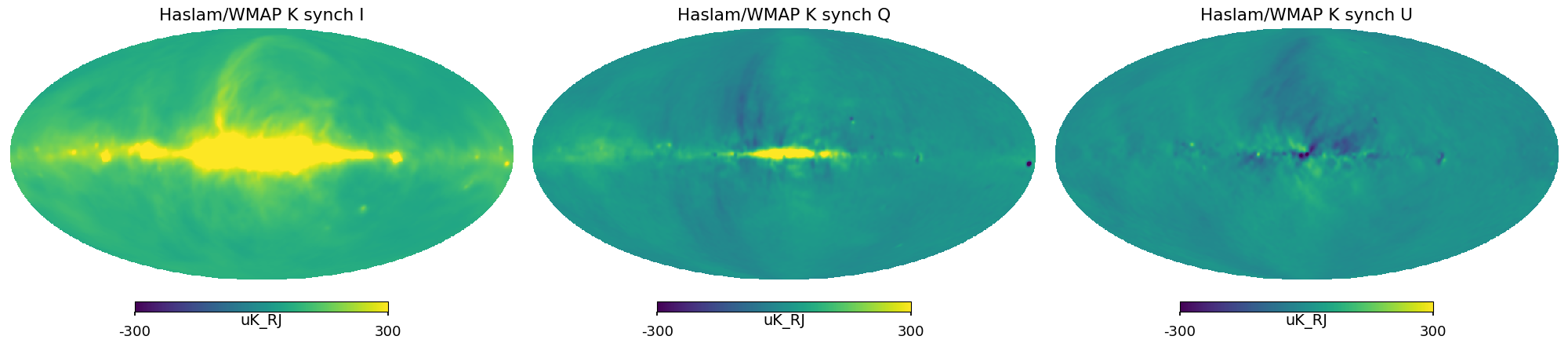
We use the Haslam map at 408MHz for the intensity template
for Q and U we adopt maps from the K-band of WMAP .
rescaling Haslam map to 23 GHz assuming a constant spectral index =-3.1 and from K to uK
converting Wmap maps to from mK to uK
we reduce noise by smoothing to 2 deg
[16]:
rescaling_factor = (23.0 / 0.408) ** -3.1
imap *= rescaling_factor
imap <<= u.K_RJ
imap = imap.to(u.uK_RJ)
qumap <<= u.mK_RJ
qumap = qumap.to("uK_RJ")
[17]:
IQU = np.array([imap, qumap[0], qumap[1]])
[18]:
FWHM_SMOOTHING = 2 # deg
IQU = hp.smoothing(IQU, fwhm=np.radians(FWHM_SMOOTHING))
IQU <<= u.uK_RJ
[19]:
plt.figure(figsize=(20, 5))
for i_pol, pol in components:
hp.mollview(
IQU[i_pol],
title="Haslam/WMAP K synch " + pol,
sub=131 + i_pol,
unit=IQU.unit,
min=-300,
max=300,
)
[20]:
if not os.path.exists(datadir / "synch_hybrid.fits"):
hp.write_map(datadir / "synch_hybrid.fits", IQU)
[21]:
import numpy as np
import healpy as hp
def map_to_log_pol_tens(m):
P = np.sqrt(m[1] ** 2 + m[2] ** 2)
log_pol_tens = np.empty_like(m)
log_pol_tens[0] = np.log(m[0] ** 2 - P**2) / 2.0
log_pol_tens[1:] = m[1:] / P * np.log((m[0] + P) / (m[0] - P)) / 2.0
return log_pol_tens
def log_pol_tens_to_map(log_pol_tens):
P = np.sqrt(log_pol_tens[1] ** 2 + log_pol_tens[2] ** 2)
m = np.empty_like(log_pol_tens)
exp_i = np.exp(log_pol_tens[0])
m[0] = exp_i * np.cosh(P)
m[1:] = log_pol_tens[1:] / P * exp_i * np.sinh(P)
return m
def sigmoid(x, x0, width, power=4):
"""Sigmoid function given start point and width
Parameters
----------
x : array
input x axis
x0 : float
value of x where the sigmoid starts (not the center)
width : float
width of the transition region in unit of x
power : float
tweak the steepness of the curve
Returns
-------
sigmoid : array
sigmoid, same length of x"""
return 1.0 / (1 + np.exp(-power * (x - x0 - width / 2) / width))
[22]:
iqu = map_to_log_pol_tens(IQU.value)
/tmp/ipykernel_17579/1699011499.py:8: RuntimeWarning: invalid value encountered in log
log_pol_tens[0] = np.log(m[0] ** 2 - P**2) / 2.0
/tmp/ipykernel_17579/1699011499.py:9: RuntimeWarning: invalid value encountered in log
log_pol_tens[1:] = m[1:] / P * np.log((m[0] + P) / (m[0] - P)) / 2.0
[23]:
m_back = log_pol_tens_to_map(iqu)
hp.mollview((IQU.value - m_back)[1], min=-1e-12, max=1e-12)
del m_back
[24]:
print(
f"{np.isnan(iqu[0]).sum() } pixels out of { iqu[0].size} are NaNs in Log Pol Tens maps "
)
for i in range(3):
iqu[i, np.isnan(iqu[i])] = np.nanmedian(iqu[i])
assert np.isnan(iqu).sum() == 0
print(
f"{np.isnan(iqu[0]).sum() } pixels out of { iqu[0].size} are NaNs in Log Pol Tens maps "
)
527 pixels out of 3145728 are NaNs in Log Pol Tens maps
0 pixels out of 3145728 are NaNs in Log Pol Tens maps
[25]:
if not os.path.exists(datadir / "synch_logpoltens.fits"):
hp.write_map(datadir / "synch_logpoltens.fits", iqu)
[26]:
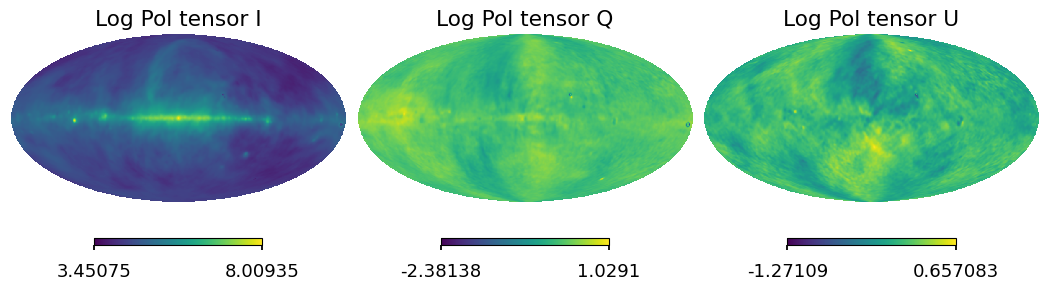
for i_pol, pol in components:
hp.mollview(iqu[i_pol], title="Log Pol tensor " + pol, sub=131 + i_pol)
[27]:
from scipy.optimize import curve_fit
[28]:
def model(ell, A, gamma):
out = A * ell**gamma
return out
[29]:
def run_anafast(m, lmax):
clanaf = hp.anafast(m, lmax=lmax)
cl = {}
cl["TT"] = clanaf[0]
cl["EE"] = clanaf[1]
cl["BB"] = clanaf[2]
cl["TE"] = clanaf[3]
ell = np.arange(lmax + 1, dtype=float)
cl_norm = ell * (ell + 1) / np.pi / 2
cl_norm[0] = 1
return ell, cl_norm, cl
def run_namaster(m, mask, lmax, nlbins):
"""Compute C_ell with NaMaster
Parameters
----------
m : numpy array
T only or TQU HEALPix map
mask : numpy array
mask, 1D, 0 for masked pixels,
needs to have same Nside of the input map
lmax : int
maximum ell of the spherical harmonics transform
Returns
-------
ell : numpy array
array of ell from 0 to lmax (length lmax+1)
cl_norm : numpy array
ell (ell+1)/2pi factor to turn C_ell into D_ell
first element is set to 1
cl : dict of numpy arrays
dictionary of numpy arrays with all components
of the spectra, for now only II, EE, BB, no
cross-spectra
"""
nside = hp.npix2nside(len(mask))
# b = nmt.NmtBin.from_nside_linear(nside, 16)
# leff = b.get_effective_ells()
binning = nmt.NmtBin(nside=nside, nlb=nlbins, lmax=lmax, is_Dell=False)
cl = {}
if len(m) == 3:
f_0 = nmt.NmtField(mask, [m[0]])
f_2 = nmt.NmtField(
mask, m[1:].copy(), purify_b=True
) # NaMaster masks the map in-place
cl_namaster = nmt.compute_full_master(f_2, f_2, binning)
cl["EE"] = np.concatenate([[0, 0], cl_namaster[0]])
cl["BB"] = np.concatenate([[0, 0], cl_namaster[3]])
cl_namaster = nmt.compute_full_master(f_0, f_2, binning)
cl["TE"] = np.concatenate([[0, 0], cl_namaster[0]])
elif m.ndim == 1:
m = m.reshape((1, -1))
f_0 = nmt.NmtField(mask, [m[0]])
cl_namaster_I = nmt.compute_full_master(f_0, f_0, binning)
cl["TT"] = np.concatenate([[0, 0], cl_namaster_I[0]])
ell = np.concatenate([[0, 1], binning.get_effective_ells()])
cl_norm = ell * (ell + 1) / np.pi / 2
cl_norm[0] = 1
return ell, cl_norm, cl
[30]:
ell, cl_norm, cl = run_anafast(iqu, lmax)
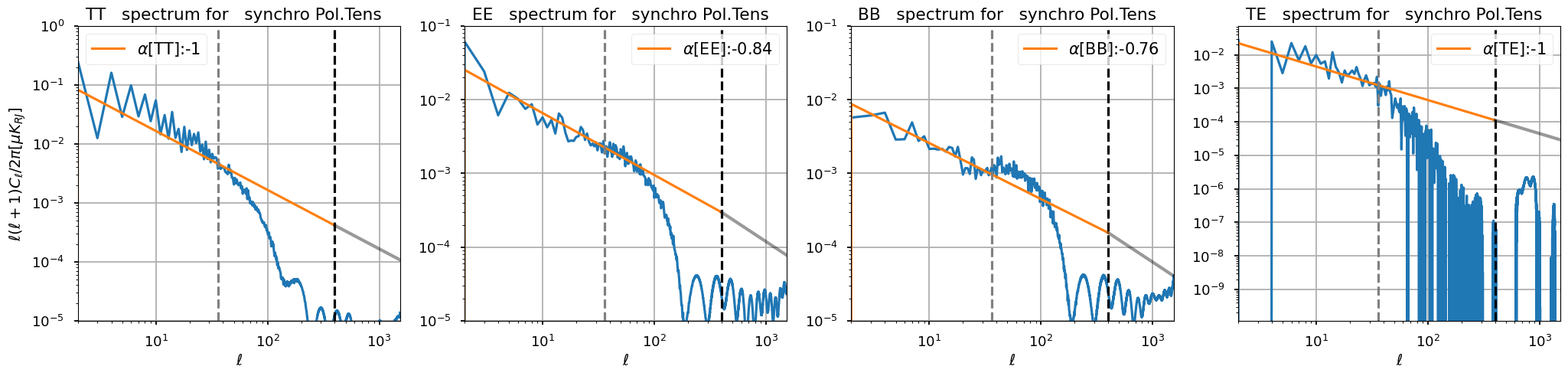
Power law fit¶
We fit in\(\ell \in[1-,36]\) a power law for TT spectrum of the logpol tens i map
we use the spectral index power law also for EE and BB power spectra to avoid unphysical crossings of EE over TT and BB over TT at higher multipoles
[31]:
ell_fit_low = {"TT": 10, "EE": 10, "BB": 10, "TE": 10}
ell_fit_high = {"TT": 36, "EE": 36, "BB": 36, "TE": 36}
gamma_fit2 = {"TT": -1, "EE": -0.84, "BB": -0.76, "TE": -1}
A_fit, gamma_fit, A_fit_std, gamma_fit_std = {}, {}, {}, {}
plt.figure(figsize=(25, 5))
A_fit2 = {}
smallscales = []
ell_pivot = 400
for ii, pol in enumerate(spectra_components):
plt.subplot(141 + ii)
xdata = np.arange(ell_fit_low[pol], ell_fit_high[pol])
ydata = xdata * (xdata + 1) / np.pi / 2 * cl[pol][xdata]
(A_fit[pol], gamma_fit[pol]), cov = curve_fit(model, xdata, ydata)
A_fit2[pol] = np.fabs(A_fit[pol]) * ell_fit_high[pol] ** (
gamma_fit[pol] - gamma_fit2[pol]
)
plt.loglog(ell, ell * (ell + 1) / np.pi / 2 * cl[pol])
scaling = model(ell[:ell_pivot], A_fit2[pol], gamma_fit2[pol])
scaling[:2] = 0
plt.plot(ell[:ell_pivot], scaling, label=r"$\alpha$" + f"[{pol}]:{gamma_fit2[pol]}")
smallscales.append(scaling)
plt.axvline(ell_fit_high[pol], linestyle="--", color="gray")
plt.axvline(ell_pivot, linestyle="--", color="k")
plt.grid()
plt.title(f"{pol} spectrum for synchro Pol.Tens ")
plt.xlabel(("$\ell$"))
plt.xlim(2, lmax)
plt.legend(fontsize=15)
for ii, pol in enumerate(spectra_components):
# we change the EE and BB power laws
xdata = np.arange(ell_fit_high[pol], ell.size)
ydata = xdata * (xdata + 1) / np.pi / 2 * cl[pol][xdata]
(A_fit[pol], gamma_fit[pol]), cov = curve_fit(model, xdata, ydata)
plt.subplot(141 + ii)
if pol == "TE":
A_fit2[pol] = A_fit2[pol] * ell_pivot ** (gamma_fit2[pol] - gamma_fit2["TE"])
scaling = model(ell[ell_pivot:], A_fit2[pol], gamma_fit2["TE"])
plt.plot(
ell[ell_pivot:],
scaling,
linewidth=3,
alpha=0.4,
color="k",
)
smallscales[ii] = np.concatenate([smallscales[ii], scaling])
else:
A_fit2[pol] = A_fit2[pol] * ell_pivot ** (gamma_fit2[pol] - gamma_fit2["TT"])
scaling = model(ell[ell_pivot:], A_fit2[pol], gamma_fit2["TT"])
plt.plot(
ell[ell_pivot:],
scaling,
linewidth=3,
alpha=0.4,
color="k",
)
smallscales[ii] = np.concatenate([smallscales[ii], scaling])
plt.subplot(141)
plt.ylabel("$\ell(\ell+1)C_\ell/2\pi [\mu K_{RJ}]$")
plt.ylim(1e-5, 1e0)
plt.subplot(142)
plt.ylim(1e-5, 1e-1)
plt.subplot(143)
plt.ylim(1e-5, 1e-1)
plt.subplot(143)
plt.ylim(1e-5, 1e-1)
/tmp/ipykernel_17579/3875259470.py:2: RuntimeWarning: divide by zero encountered in reciprocal
out = A * ell**gamma
/tmp/ipykernel_17579/3875259470.py:2: RuntimeWarning: divide by zero encountered in power
out = A * ell**gamma
[31]:
(1e-05, 0.1)
[32]:
cl_ss = [
smallscales[ii] * sigmoid(ell, ell_fit_high[pol], ell_fit_high[pol] / 10) / cl_norm
for ii, pol in enumerate(spectra_components)
]
(new) modulation for intensity and polarization¶
downgrade map at
nside=128estimate power spectra masking the galactic plane
[33]:
import pymaster as nmt
mask = IQU[0].value > 150
iqu2 = hp.ud_grade(
iqu, # hp.smoothing(iqu, fwhm=np.radians(np.sqrt(3**2- FWHM_SMOOTHING**2) ) )
nside_out=128,
)
mask = hp.ud_grade(mask, nside_out=hp.get_nside(iqu2))
lmax_mod = 100
apo_mask = nmt.mask_apodization(mask, 5, apotype="C2")
ell0, norm, cl0 = run_anafast(iqu2 * mask, lmax=lmax_mod)
[34]:
nsidepatches = 8
centers = np.vstack(
hp.pix2vec(ipix=np.arange(hp.nside2npix(nsidepatches)), nside=nsidepatches)
).T
def bin_cell(cell, dig):
cb = []
errb = []
for i in np.unique(dig):
msk = dig == i
cb.append(cell[msk].mean())
errb.append(cell[msk].std())
return np.array(cb), np.array(errb)
def bin_ell(ells, dig):
lb = []
dl = []
for i in np.unique(dig):
msk = dig == i
lb.append(ells[msk].mean())
dl.append((ells[msk].max() - ells[msk].min()) / 2)
return np.array(lb), np.array(dl)
fit_model = lambda x, Ad, alpha: Ad * (x / 30) ** (alpha)
tmod = np.zeros_like(iqu2[0])
pmod = np.zeros_like(iqu2[0])
n_eff = np.zeros_like(iqu2[0])
for ipix, c in enumerate(centers):
patch = np.zeros_like(iqu2[0])
maskpixs = hp.query_disc(nside=hp.get_nside(iqu2), vec=c, radius=np.radians(15.6))
patch[maskpixs] = 1
apo_patch = nmt.mask_apodization(patch, 5, apotype="C2")
fsky = apo_patch.sum() / apo_patch.size
if ipix % 100 == 0:
print(ipix)
ellp, norm, clp = run_anafast(m=iqu2 * apo_patch, lmax=lmax_mod)
digi = np.digitize(ellp, np.linspace(0, lmax_mod, 10))
dtt, errtt = bin_cell((clp["TT"]), digi) / fsky
dee, erree = bin_cell((clp["EE"]), digi) / fsky
lb, delta_l = bin_ell(ellp, digi)
lmask = np.logical_and((lb) < 50, lb > 10)
lmaskt = np.logical_and((lb) < 50, lb > 10)
param_ee, _ = curve_fit(
fit_model, ydata=dee[lmask], xdata=lb[lmask], sigma=erree[lmask]
)
param_tt, _ = curve_fit(
fit_model, ydata=dtt[lmaskt], xdata=lb[lmaskt], sigma=errtt[lmaskt]
)
l_ = 36
pmod += np.sqrt(fit_model(l_, *param_ee) / cl0["EE"][l_]) * apo_patch
tmod += np.sqrt(fit_model(l_, *param_tt) / cl0["TT"][l_]) * apo_patch
n_eff += apo_patch
# break
0
100
200
300
400
500
600
700
[35]:
psm = hp.smoothing(pmod / n_eff, fwhm=np.radians(11.5))
tsm = hp.smoothing(tmod / n_eff, fwhm=np.radians(11.5))
hp.mollview(psm, sub=122, title="P Modulation")
hp.mollview(tsm, sub=121, title="I Modulation")
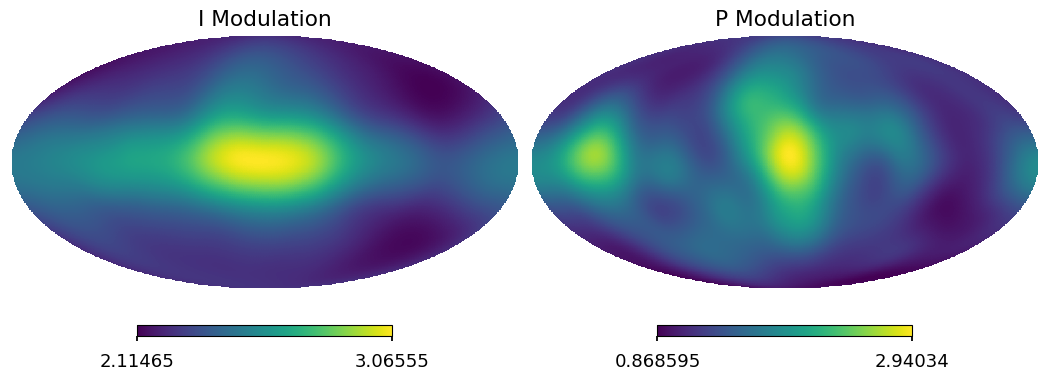
[36]:
LS_lmax = 128
[37]:
np.random.seed(777)
# filter large scales
alm_log_pol_tens_fullsky = hp.map2alm(iqu, lmax=lmax, use_pixel_weights=True)
ii_LS_alm = np.empty_like(alm_log_pol_tens_fullsky)
for ii, pol in enumerate(spectra_components[:-1]):
sig_func = sigmoid(ell, x0=ell_fit_high[pol], width=ell_fit_high[pol] / 10)
ii_LS_alm[ii] = hp.almxfl(alm_log_pol_tens_fullsky[ii], (1.0 - sig_func) ** 0.2)
[38]:
for pol in range(3):
plt.loglog(hp.alm2cl(ii_LS_alm[pol]))
plt.grid()
plt.axvline(LS_lmax, color="grey", linestyle="--");
[39]:
ii_LS_alm = hp.resize_alm(ii_LS_alm, lmax, lmax, LS_lmax, LS_lmax)
log_ss = hp.synfast(
cl_ss,
lmax=lmax,
new=True,
nside=hp.get_nside(iqu),
)
log_ss_modulated = np.zeros_like(log_ss)
log_ss_modulated[0] = log_ss[0] * (
hp.ud_grade(tsm / tsm.max(), nside_out=hp.get_nside(log_ss))
) # modulate_amp
log_ss_modulated[1:] = log_ss[1:] * (
hp.ud_grade(psm, nside_out=hp.get_nside(log_ss)) - 0.5
) # ampl_smooth_minmax
log_ls = hp.alm2map(ii_LS_alm, nside=nside)
ii_map_out = log_ss_modulated + log_ls
[40]:
filename = proddir / f"synch_largescale_template_logpoltens_alm_lmax{LS_lmax:d}_{version}.fits"
hp.write_alm(
filename,
ii_LS_alm,
lmax=LS_lmax,
out_dtype=np.complex64,
overwrite=True,
)
output_files.append(filename)
[41]:
pysm.utils.add_metadata(
[ filename ],
coord="G",
unit="uK_RJ",
)
[42]:
plt.figure(figsize=(15, 5))
hp.gnomview(
log_ss[0],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=152,
title="small-scales poltens i ",
)
hp.gnomview(
log_ls[0],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=153,
title="large-scales poltens i ",
)
hp.gnomview(
ii_map_out[0], reso=3.75, xsize=320, rot=[30, -58], sub=151, title="coadded i "
)
hp.gnomview(iqu[0], reso=3.75, xsize=320, rot=[30, -58], sub=155, title="input i ")
hp.gnomview(tsm, reso=3.75, xsize=320, rot=[30, -58], sub=154, title="modulation i ")
# hp.gnomview(modulate_amp, reso=3.75, xsize=320, rot=[30,-58] ,sub=154,title='modulation i ' )
plt.figure(figsize=(15, 5))
hp.gnomview(
log_ss[1],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=152,
title="small-scales poltens q",
)
hp.gnomview(
log_ls[1],
reso=3.75,
xsize=320,
rot=[30, -58],
sub=153,
title="large-scales poltens q",
)
hp.gnomview(
ii_map_out[1], reso=3.75, xsize=320, rot=[30, -58], sub=151, title="coadded q "
)
hp.gnomview(iqu[1], reso=3.75, xsize=320, rot=[30, -58], sub=155, title="input q ")
hp.gnomview(psm, reso=3.75, xsize=320, rot=[30, -58], sub=154, title="modulation p ")
# hp.gnomview(ampl_smooth_minmax , reso=3.75, xsize=320, rot=[30,-58] ,sub=154,title='modulation p ' )
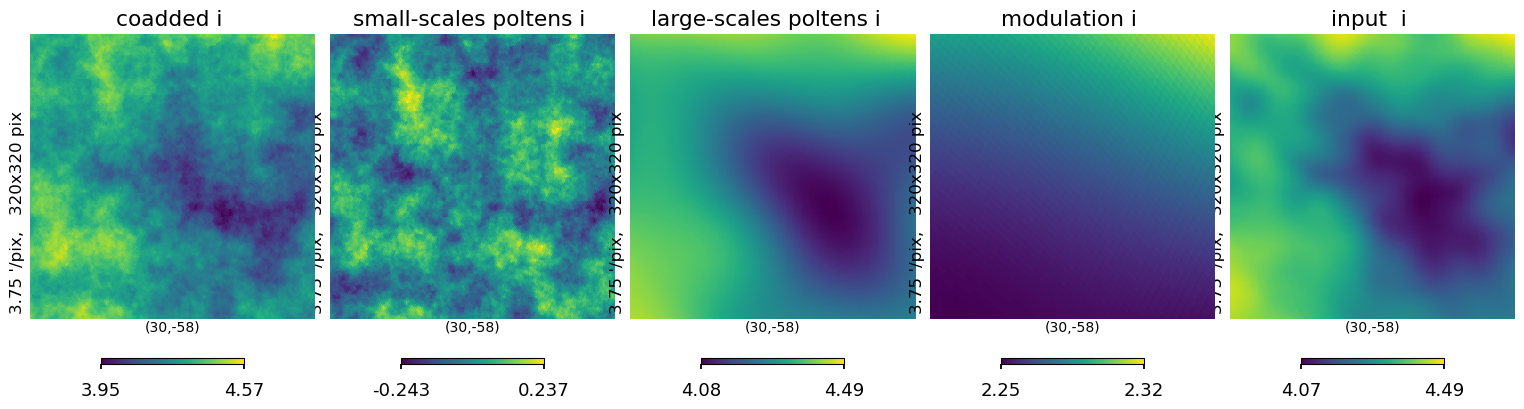
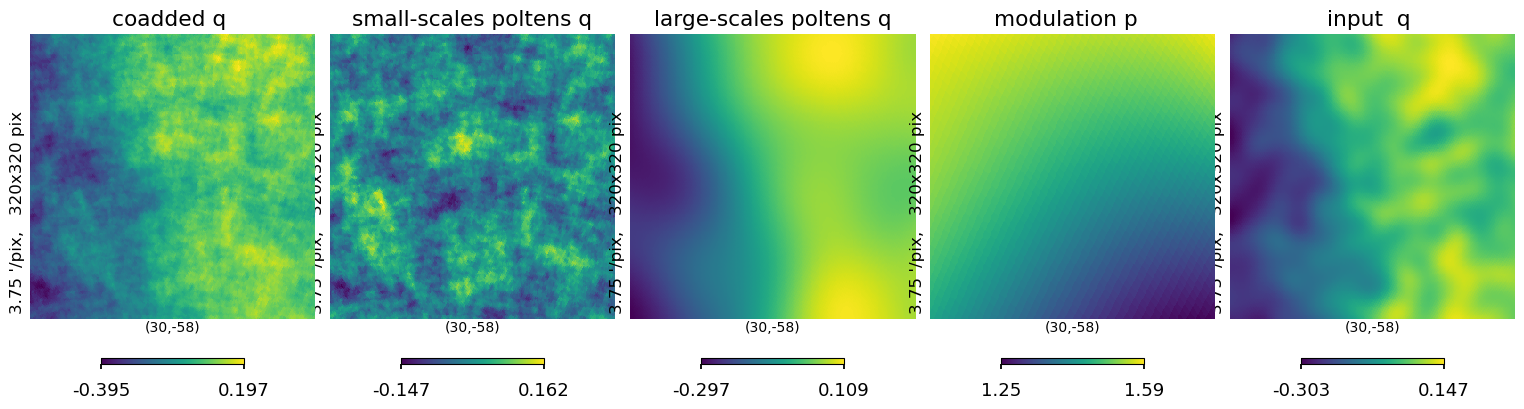
[43]:
ell, cl_norm, cltot = run_anafast(ii_map_out, lmax)
[44]:
plt.figure(figsize=(25, 5))
for ii, pol in enumerate(spectra_components):
plt.subplot(141 + ii)
plt.loglog(ell, ell * (ell + 1) / np.pi / 2 * cl[pol], label="anafast $C_\ell$")
plt.loglog(ell, ell * (ell + 1) / np.pi / 2 * cltot[pol], label="tot $C_\ell$")
# plt.loglog(ell, ell*(ell+1)/np.pi/2 * small_scales_input_cl[ii] )
plt.axvline(
ell_fit_low[pol],
linestyle="--",
color="black",
label="$ \ell={} $".format(ell_fit_low[pol]),
)
plt.axvline(
ell_fit_high[pol],
linestyle="--",
color="gray",
label="$ \ell={} $".format(ell_fit_high[pol]),
)
plt.legend()
plt.grid()
plt.title(
f"{pol} spectrum for synch Pol.Tens "
+ r"$\gamma_{fit}=$"
+ f"{gamma_fit[pol]:.2f}"
)
plt.ylabel("$\ell(\ell+1)C_\ell/2\pi [\mu K_{RJ}]$")
plt.xlabel(("$\ell$"))
plt.xlim(2, lmax)
# plt.ylim(1e-5, 1e-1)
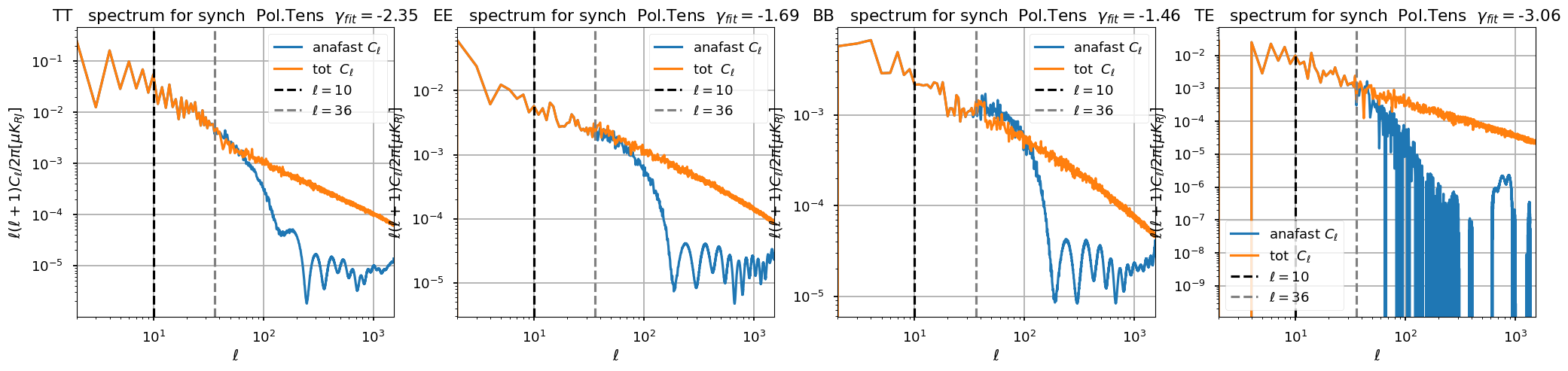
[45]:
galplane = IQU[0].value > 200
galplane_apo = nmt.mask_apodization(galplane, 5, apotype="C2")
[46]:
output_map = log_pol_tens_to_map(ii_map_out)
output_map = output_map * (1 - galplane_apo) + IQU.value * galplane_apo
[47]:
hp.mollview(output_map[0], min=0, max=500)
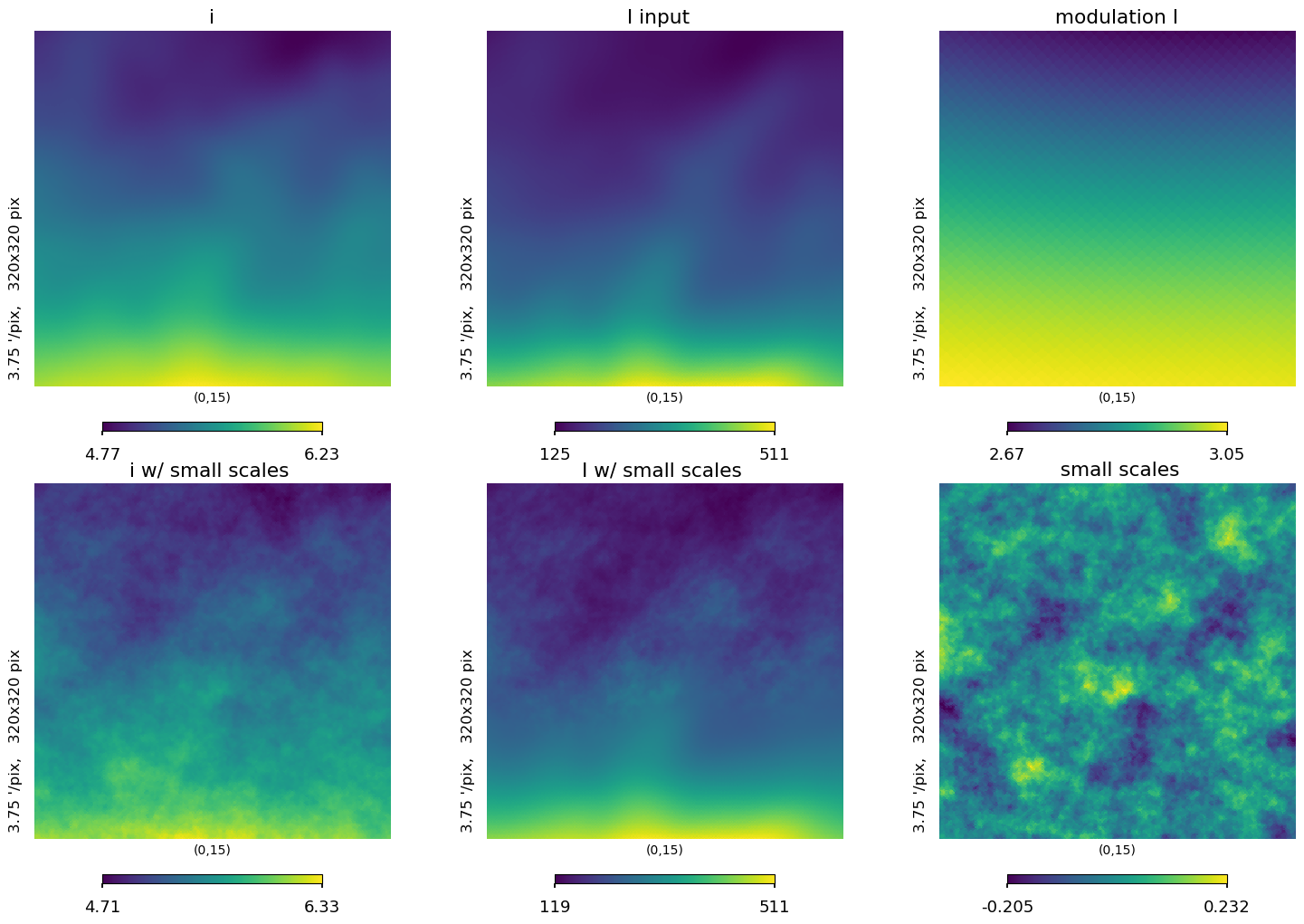
[48]:
lat = 15
plt.figure(figsize=(15, 10))
hp.gnomview(
ii_map_out[0],
title="i w/ small scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=234,
)
hp.gnomview(iqu[0], title="i", rot=[0, lat], reso=3.75, xsize=320, sub=231)
hp.gnomview(
tsm,
title=" modulation I ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=233,
)
hp.gnomview(
(IQU[0]),
title=" I input ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=232,
)
hp.gnomview(
(log_ss_modulated)[0],
title=" small scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=236,
)
hp.gnomview(
output_map[0],
title="I w/ small scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=235,
)
lat = 15
plt.figure(figsize=(15, 10))
hp.gnomview(
ii_map_out[1],
title="q w/ small scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=234,
)
hp.gnomview(iqu[1], title="q", rot=[0, lat], reso=3.75, xsize=320, sub=231)
hp.gnomview(
(psm),
title=" modulation P ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=233,
)
hp.gnomview(
(IQU[1]),
title=" Q input ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=232,
)
hp.gnomview(
(log_ss_modulated)[1],
title=" small scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=236,
)
hp.gnomview(
output_map[1],
title="Q w/ small scales ",
rot=[0, lat],
reso=3.75,
xsize=320,
sub=235,
)
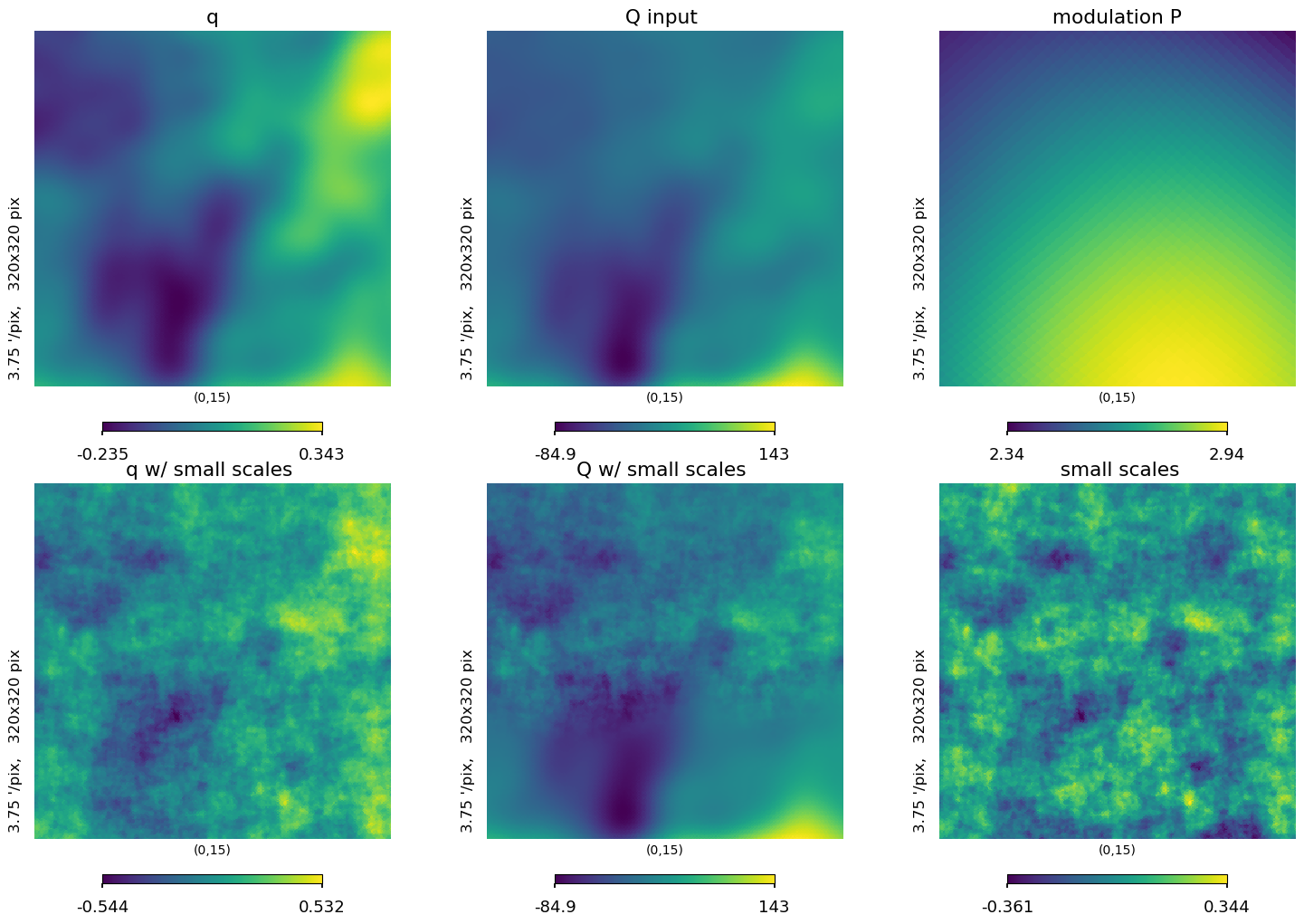
[49]:
if not os.path.exists(datadir / "sync_pysm_mod_out_circles.fits"):
hp.write_map(datadir / "sync_pysm_mod_out_circles.fits", output_map, overwrite=True)
[50]:
bkmaskfile = datadir / "bk14_mask_gal_n0512.fits"
if not bkmaskfile.exists():
!wget -O $bkmaskfile http://bicepkeck.org/BK14_datarelease/bk14_mask_gal_n0512.fits
maskbk = hp.read_map(bkmaskfile, verbose=False)
idx = np.where((maskbk < 0) | (~np.isfinite(maskbk)))
maskbk[idx] = 0
WARNING: AstropyDeprecationWarning: "verbose" was deprecated in version 1.15.0 and will be removed in a future version. [warnings]
[51]:
aoa = "/global/cscratch1/sd/flauger/S4_misc/weights_aoa_spsat_apo_gal_512.fits"
mask_aoa = hp.read_map(aoa, verbose=False)
WARNING: AstropyDeprecationWarning: "verbose" was deprecated in version 1.15.0 and will be removed in a future version. [warnings]
[52]:
planck_mask_filename = datadir / "HFI_Mask_GalPlane-apo2_2048_R2.00.fits"
if not planck_mask_filename.exists():
!wget -O $planck_mask_filename "http://pla.esac.esa.int/pla/aio/product-action?MAP.MAP_ID=HFI_Mask_GalPlane-apo2_2048_R2.00.fits"
[53]:
planck_masks = {
"GAL099": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL099"]), nside_out=512),
"GAL097": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL097"]), nside_out=512),
"GAL090": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL090"]), nside_out=512),
"GAL080": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL080"]), nside_out=512),
"GAL070": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL070"]), nside_out=512),
"GAL060": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL060"]), nside_out=512),
"GAL040": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL040"]), nside_out=512),
"GAL020": hp.ud_grade(hp.read_map(planck_mask_filename, ["GAL020"]), nside_out=512),
}
[54]:
ell, cl_norm, clo = run_anafast(output_map, lmax=2048)
ell, cl_norm, cli = run_anafast(IQU, lmax=2048)
[55]:
colors = plt.cm.cividis(np.linspace(0, 1, len(planck_masks.keys()) + 1))
[56]:
bins = {
"GAL099": 5,
"GAL097": 5,
"GAL090": 5,
"GAL080": 5,
"GAL070": 15,
"GAL060": 15,
"GAL040": 25,
"GAL020": 25,
}
plt.figure(figsize=(15, 10))
import os
# for jj, k in enumerate([ "GAL080","GAL040","GAL020" ]) :
for jj, k in enumerate(planck_masks.keys()):
print(k)
fspectra = datadir / f"sync_input_gal{k[3:]}_spectra.npz"
if os.path.exists(fspectra):
input_ell = np.load(fspectra)["ell"]
cl_in = {kk: np.load(fspectra)[kk] for kk in spectra_components + ["TE"]}
else:
input_ell, cl_norm, cl_in = run_namaster(
m=IQU, mask=planck_masks[k], lmax=1300, nlbins=bins[k]
)
np.savez(
fspectra,
ell=input_ell,
TT=cl_in["TT"],
EE=cl_in["EE"],
BB=cl_in["BB"],
TE=cl_in["TE"],
)
fspectra = datadir / f"sync_galplane_gal{k[3:]}_spectra.npz"
if os.path.exists(fspectra):
output_ell = np.load(fspectra)["ell"]
cl_out = {kk: np.load(fspectra)[kk] for kk in spectra_components + ["TE"]}
else:
output_ell, cl_norm, cl_out = run_namaster(
m=output_map, mask=planck_masks[k], lmax=1300, nlbins=bins[k]
)
np.savez(
fspectra,
ell=output_ell,
TT=cl_out["TT"],
EE=cl_out["EE"],
BB=cl_out["BB"],
TE=cl_out["TE"],
)
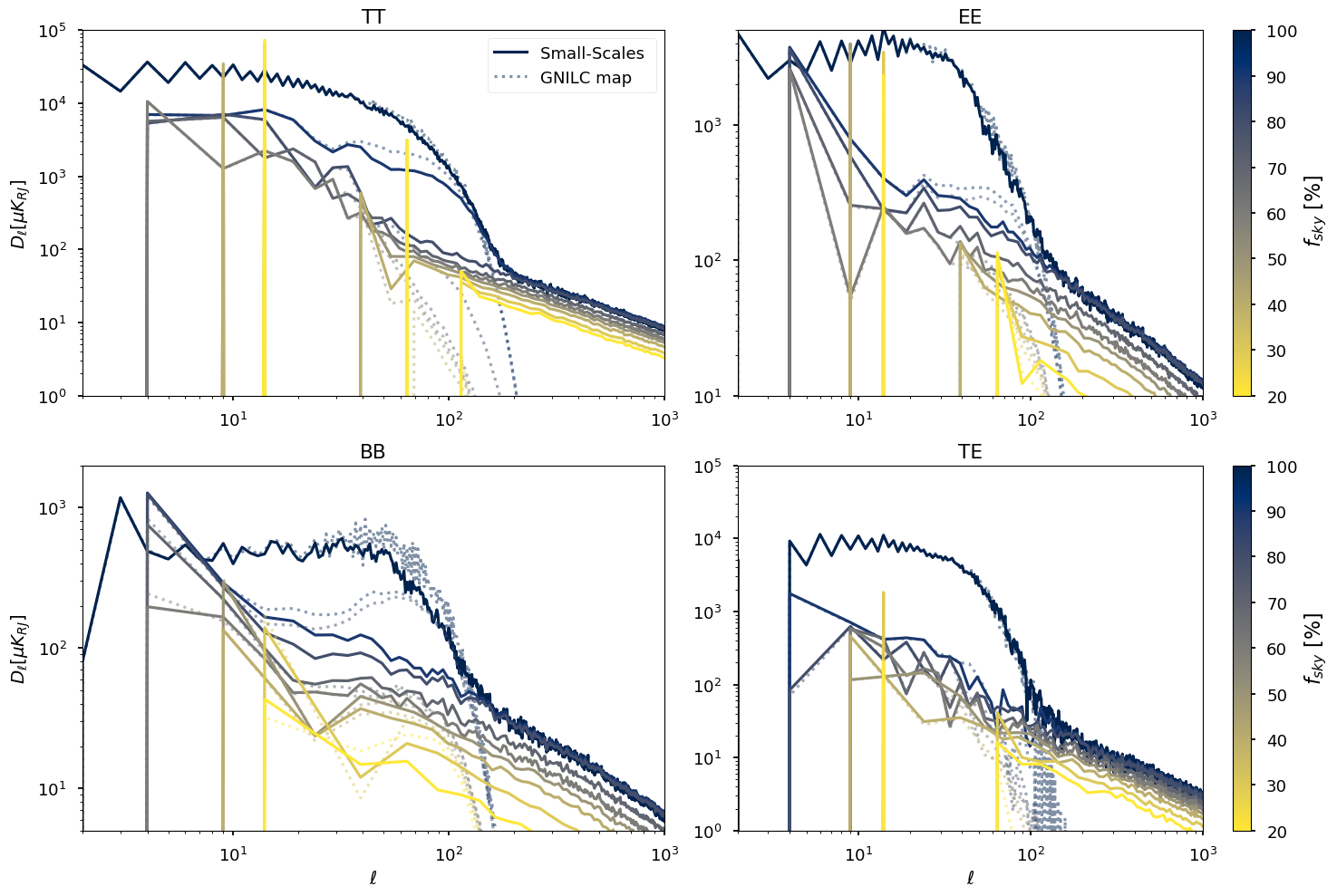
for ii, pol in enumerate(["TT", "EE", "BB", "TE"]):
plt.subplot(2, 2, ii + 1)
if jj == 0:
plt.title(pol)
plt.loglog(
ell,
ell**2 * (clo[pol]),
label="Small-Scales ",
color=colors[0],
)
plt.loglog(
ell,
ell**2 * (cli[pol]),
":",
label="GNILC map ",
color=colors[0],
alpha=0.5,
)
plt.loglog(output_ell, output_ell**2 * (cl_out[pol]), color=colors[jj + 1])
# ,label =f"{k}")
plt.loglog(
input_ell,
input_ell**2 * (cl_in[pol]),
":",
color=colors[jj + 1],
alpha=0.5,
)
plt.subplot(221)
plt.legend()
plt.ylabel("$ D_\ell [\mu K_{RJ}]$")
plt.ylim(1e0, 1e5)
plt.xlim(2, 1e3)
plt.subplot(222)
plt.ylim(1e1, 5e3)
plt.xlim(2, 1e3)
sm = plt.cm.ScalarMappable(
cmap=plt.cm.cividis_r, norm=plt.Normalize(vmin=20.0, vmax=100)
)
cb = plt.colorbar(sm)
cb.set_label(r" $f_{sky}$ [%]", rotation=90, fontsize=16)
plt.subplot(223)
plt.ylabel("$ D_\ell [\mu K_{RJ}]$")
plt.xlabel(("$\ell$"))
plt.ylim(5e0, 2e3)
plt.xlim(2, 1e3)
plt.subplot(224)
plt.ylim(1e0, 1e5)
plt.xlim(2, 1e3)
plt.xlabel(("$\ell$"))
sm = plt.cm.ScalarMappable(
cmap=plt.cm.cividis_r, norm=plt.Normalize(vmin=20.0, vmax=100)
)
cb = plt.colorbar(sm)
cb.set_label(r"$f_{sky}$ [%]", rotation=90, fontsize=16)
plt.tight_layout()
GAL099
GAL097
GAL090
GAL080
GAL070
GAL060
GAL040
GAL020
/tmp/ipykernel_17579/791073269.py:92: MatplotlibDeprecationWarning: Unable to determine Axes to steal space for Colorbar. Using gca(), but will raise in the future. Either provide the *cax* argument to use as the Axes for the Colorbar, provide the *ax* argument to steal space from it, or add *mappable* to an Axes.
cb = plt.colorbar(sm)
/tmp/ipykernel_17579/791073269.py:109: MatplotlibDeprecationWarning: Unable to determine Axes to steal space for Colorbar. Using gca(), but will raise in the future. Either provide the *cax* argument to use as the Axes for the Colorbar, provide the *ax* argument to steal space from it, or add *mappable* to an Axes.
cb = plt.colorbar(sm)
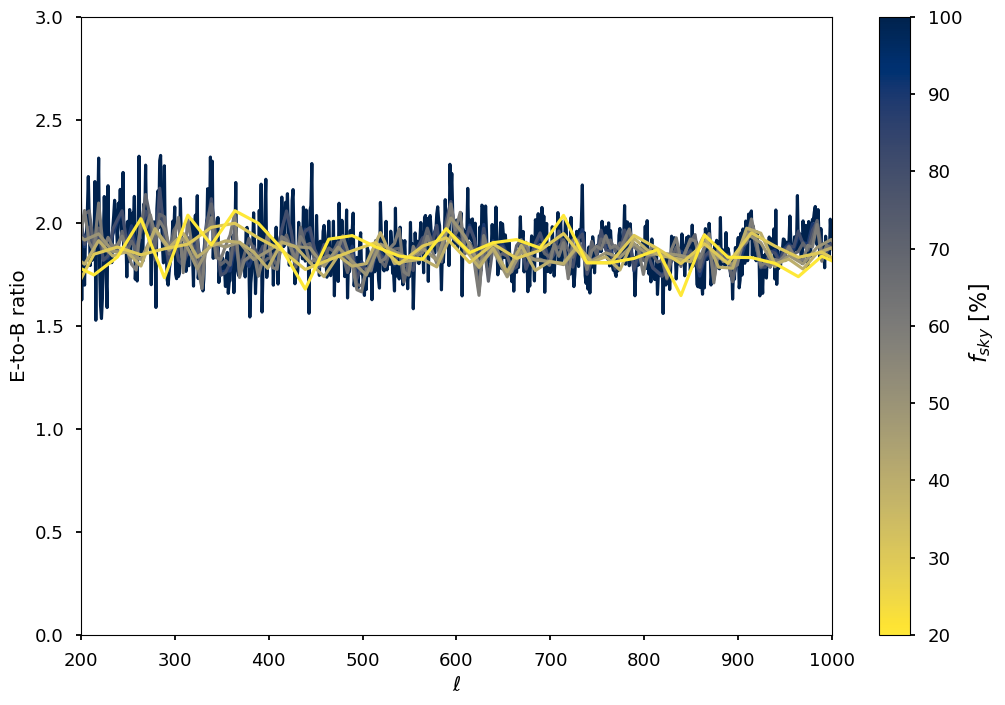
[57]:
# plt.figure(figsize=(15,10 ))
import os
for jj, k in enumerate(planck_masks.keys()):
fspectra = datadir / f"sync_galplane_gal{k[3:]}_spectra.npz"
if os.path.exists(fspectra):
# print("read Namaster spectra ")
output_ell = np.load(fspectra)["ell"]
cl_out = {kk: np.load(fspectra)[kk] for kk in spectra_components + ["TE"]}
if jj == 0:
plt.plot(ell, (clo["EE"] / clo["BB"]), color=colors[0], alpha=1)
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
plt.xlim(200, 1e3)
plt.ylim(0, 3)
sm = plt.cm.ScalarMappable(
cmap=plt.cm.cividis_r, norm=plt.Normalize(vmin=20.0, vmax=100)
)
cb = plt.colorbar(sm)
cb.set_label(r" $f_{sky}$ [%]", rotation=90, fontsize=16)
plt.xlabel(("$\ell$"))
plt.ylabel(("E-to-B ratio"))
plt.tight_layout()
/tmp/ipykernel_17579/972234778.py:13: RuntimeWarning: invalid value encountered in divide
plt.plot(ell, (clo["EE"] / clo["BB"]), color=colors[0], alpha=1)
/tmp/ipykernel_17579/972234778.py:14: RuntimeWarning: invalid value encountered in divide
plt.plot(output_ell, (cl_out["EE"] / cl_out["BB"]), color=colors[jj + 1], alpha=1)
/tmp/ipykernel_17579/972234778.py:21: MatplotlibDeprecationWarning: Unable to determine Axes to steal space for Colorbar. Using gca(), but will raise in the future. Either provide the *cax* argument to use as the Axes for the Colorbar, provide the *ax* argument to steal space from it, or add *mappable* to an Axes.
cb = plt.colorbar(sm)
[58]:
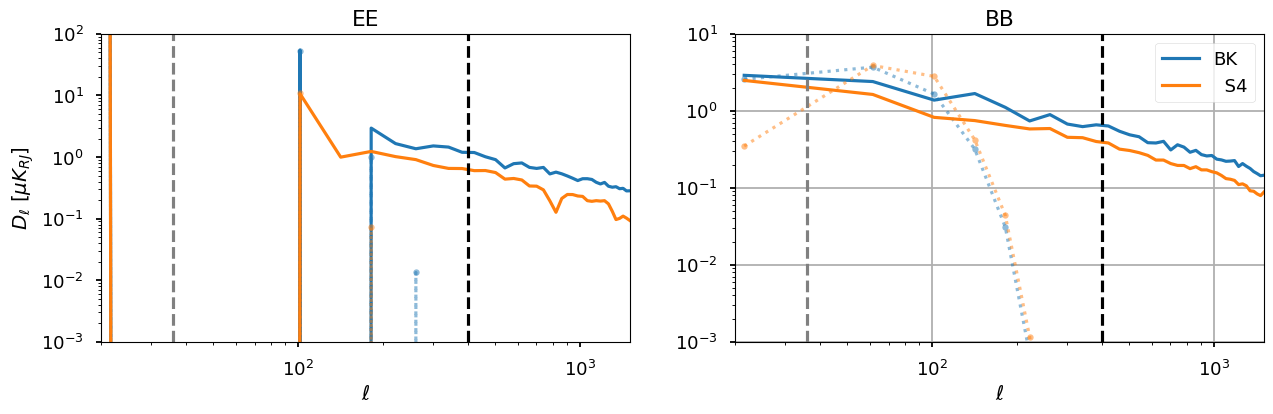
def ClBB(mask, mapp):
map1 = mapp.copy()
b = nmt.NmtBin.from_nside_linear(hp.get_nside(mask), 40)
f_2 = nmt.NmtField(mask, map1, purify_b=True)
cl_22 = nmt.compute_full_master(f_2, f_2, b)
ell_arr = b.get_effective_ells()
return (ell_arr, cl_22[3], cl_22[0])
[59]:
import matplotlib
import matplotlib.pyplot as plt
from matplotlib import rc
[60]:
i_dic = {}
(ells, clBB_i, clEE_i) = ClBB(mask_aoa, IQU[1:, :])
i_dic["S4"] = [clBB_i, clEE_i]
(ells, clBB_i, clEE_i) = ClBB(maskbk, IQU[1:, :])
i_dic["BK"] = [clBB_i, clEE_i]
[61]:
o_dic = {}
(oells, clBB_o, clEE_o) = ClBB(mask_aoa, output_map[1:, :])
o_dic["S4"] = [clBB_o, clEE_o]
(oells, clBB_o, clEE_o) = ClBB(maskbk, output_map[1:, :])
o_dic["BK"] = [clBB_o, clEE_o]
[62]:
plt.figure(figsize=(15, 4))
plt.subplot(121)
plt.title("EE")
plt.ylabel("$ D_\ell\,\, [\mu K_{RJ}]$")
plt.xlabel(("$\ell$"))
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["BK"][1] / (2.0 * np.pi),
".",
alpha=0.5,
color="C0",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["BK"][1] / (2.0 * np.pi),
label="BK",
color="C0",
)
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["S4"][1] / (2.0 * np.pi),
".",
alpha=0.5,
color="C1",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["S4"][1] / (2.0 * np.pi),
label="Southern-Hole",
color="C1",
)
plt.loglog()
plt.ylim(1e-3, 1e2)
plt.xlim(20, 1.5e3)
plt.axvline(ell_fit_high[pol], linestyle="--", color="gray")
plt.axvline(ell_pivot, linestyle="--", color="k")
plt.subplot(122)
plt.title("BB")
plt.xlabel(("$\ell$"))
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["BK"][0] / (2.0 * np.pi),
".",
alpha=0.5,
color="C0",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["BK"][0] / (2.0 * np.pi),
label="BK",
color="C0",
)
plt.plot(
ells,
ells * (ells + 1.0) * i_dic["S4"][0] / (2.0 * np.pi),
".",
alpha=0.5,
color="C1",
linestyle=":",
)
plt.plot(
oells,
oells * (oells + 1.0) * o_dic["S4"][0] / (2.0 * np.pi),
label=" S4",
color="C1",
)
plt.loglog()
plt.ylim(1e-3, 1e1)
plt.xlim(20, 1.5e3)
plt.axvline(ell_fit_high[pol], linestyle="--", color="gray")
plt.axvline(ell_pivot, linestyle="--", color="k")
plt.legend()
plt.grid()
[63]:
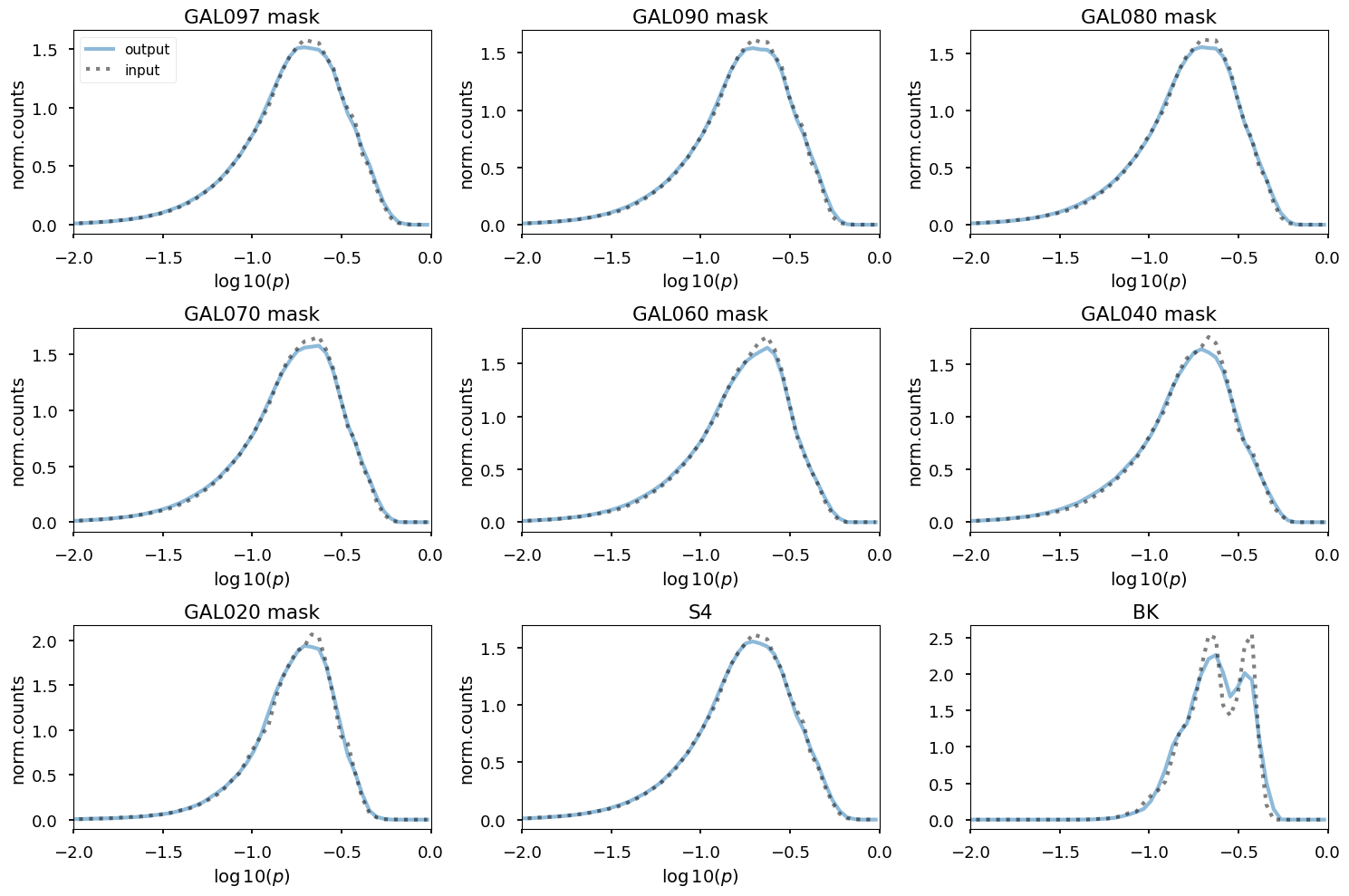
get_polfrac = lambda m: np.sqrt(m[1] ** 2 + m[2] ** 2) / m[0]
[64]:
Pout = get_polfrac(output_map)
Pin = get_polfrac(IQU.value)
logpin = np.log10(Pin)
logpout = np.log10(Pout)
[65]:
planck_masks_bool = {
k: np.ma.masked_greater(m, 0).mask for k, m in planck_masks.items()
}
[66]:
plt.figure(figsize=(15, 10))
for jj, pm in enumerate(planck_masks_bool.items()):
k = pm[0]
msk = pm[1]
print(k)
if k == "GAL099":
continue
plt.subplot(3, 3, jj)
h, edg = np.histogram(logpout[msk], bins=np.linspace(-4, 0, 100), density=True)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, color="C0", alpha=0.5, label="output")
h, edg = np.histogram(logpin[msk], density=True, bins=np.linspace(-4, 0, 100))
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, alpha=0.5, color="k", linestyle=":", label="input")
plt.ylabel("norm.counts", fontsize=14)
plt.xlabel(r"$\log10( p )$", fontsize=14)
plt.title(k + " mask")
plt.xlim(-2, -0)
plt.tight_layout()
plt.subplot(331)
plt.legend(fontsize=11, loc="upper left")
plt.subplot(339)
h, edg = np.histogram(
logpout[np.ma.masked_greater(maskbk, 0).mask],
bins=np.linspace(-4, 0, 100),
density=True,
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, color="C0", alpha=0.5, label="output")
h, edg = np.histogram(
logpin[np.ma.masked_greater(maskbk, 0).mask],
density=True,
bins=np.linspace(-4, 0, 100),
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, alpha=0.5, color="k", linestyle=":", label="input")
plt.title("BK ")
plt.xlim(-2, -0)
plt.subplot(338)
h, edg = np.histogram(
logpout[np.ma.masked_greater(mask_aoa, 0).mask],
bins=np.linspace(-4, 0, 100),
density=True,
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, color="C0", alpha=0.5, label="output")
h, edg = np.histogram(
logpin[np.ma.masked_greater(mask_aoa, 0).mask],
density=True,
bins=np.linspace(-4, 0, 100),
)
xb = np.array([(edg[i] + edg[i + 1]) / 2 for i in range(edg.size - 1)])
plt.plot(xb, h, lw=3, alpha=0.5, color="k", linestyle=":", label="input")
plt.ylabel("norm.counts", fontsize=14)
plt.xlabel(r"$\log10( p )$", fontsize=14)
plt.title("S4")
plt.xlim(-2, -0)
GAL099
GAL097
GAL090
GAL080
GAL070
GAL060
GAL040
GAL020
[66]:
(-2.0, 0.0)
Save spectra to higher ell¶
[67]:
output_nside = 8192
output_lmax = 2 * output_nside
lmax = output_lmax
ell = np.arange(output_lmax + 1)
cl_norm = ell * (ell + 1) / np.pi / 2
cl_norm[:1] = 1
[68]:
output_ell = np.arange(output_lmax + 1, dtype=np.float64)[len(smallscales[0]) :]
output_cl_norm = output_ell * (output_ell + 1) / np.pi / 2
[69]:
for ii, pol in enumerate(spectra_components):
if pol == "TE":
scaling = model(output_ell, A_fit2[pol], gamma_fit2["TE"])
smallscales[ii] = np.concatenate([smallscales[ii], scaling])
else:
scaling = model(output_ell, A_fit2[pol], gamma_fit2["TT"])
smallscales[ii] = np.concatenate([smallscales[ii], scaling])
[70]:
output_ell = np.arange(output_lmax + 1)
output_cl_norm = output_ell * (output_ell + 1) / np.pi / 2
output_cl_norm[:1] = 1
[71]:
len(smallscales[0]), len(output_cl_norm)
[71]:
(16385, 16385)
[72]:
cl_ss = [
smallscales[ii]
* sigmoid(output_ell, ell_fit_high[pol], ell_fit_high[pol] / 10)
/ output_cl_norm
for ii, pol in enumerate(spectra_components)
]
[73]:
filename = proddir / f"synch_small_scales_cl_lmax{output_lmax}_{version}.fits"
hp.write_cl(
filename,
cl_ss,
dtype=np.complex128,
overwrite=True,
)
[74]:
pysm.utils.add_metadata(
[filename],
coord="G",
unit="uK_RJ**2",
)
[75]:
output_files.append(filename)
[76]:
mod_lmax = 64
[77]:
tsm /= tsm.max()
psm -= 0.5
for name, each_modulate in [("temperature", tsm), ("polarization", psm)]:
filename = proddir / f"synch_{name}_modulation_alms_lmax{int(mod_lmax):d}_{version}.fits"
alm = hp.map2alm(each_modulate, lmax=mod_lmax, use_pixel_weights=True)
hp.write_alm(
filename,
alm,
overwrite=True,
out_dtype=np.float32,
)
pysm.utils.add_metadata(
[filename],
coord="G",
unit="",
)
output_files.append(filename)
[78]:
mod_cl_t = hp.anafast(tsm)
mod_cl_p = hp.anafast(psm)
[79]:
plt.loglog(mod_cl_t, label="mod temp")
plt.loglog(mod_cl_p, label="mod pol")
plt.axvline(mod_lmax, color="grey", linestyle="--", label="lmax")
plt.legend();
[80]:
for fitsfile in output_files:
!gzip -f $fitsfile
[ ]: